Introduction
Yunnan is located in southwest China and borders Myanmar, Laos and Vietnam. Since the first human immunodeficiency virus (HIV) epidemic in China was identified among intravenous drug users (IDUs) in Yunnan in 1989, this province has been one of the areas heavily affected by HIV-1 in China [Reference Lu1, Reference Jia2]. More importantly, Yunnan is considered as a primary gateway for the introduction of major HIV-1 subtypes into China. Thus, the changing trend of HIV-1 genetics in Yunnan is representative in China.
In the early 1990s, drug trafficking and drug abuse drove the HIV-1 epidemic in Yunnan, where subtype B and subtype C strains were introduced from Thailand [Reference Graf3, Reference Shao4] and India [Reference Luo5], respectively. In 1994, CRF01_AE was identified among female sex workers (FSWs) who had returned from Thailand to Yunnan [Reference Cheng6]. The coexistence of subtypes B and C in IDUs led to the generation of BC recombinant forms. Usually, inter-subtype recombinant genomes are common, but many of them are found in the one dually-infected individual. Such a recombinant is called a unique recombinant form (URF). If an inter-subtype recombinant genome is found in three or more persons who are not epidemiologically related, it can be classified as a circulating recombinant form (CRF), which means that it becomes one of the circulating strains in the HIV epidemic. Nowadays, it is speculated that CRF07_BC and CRF08_BC were initially established in Yunnan [Reference Feng7, Reference Tee8]. However, there was no systematic HIV-1 molecular epidemiological study carried out in the 1990s.
In the early 2000s, there were three major groups of HIV-1 circulating in Yunnan: C/CRF07_BC/CRF08_BC, CRF01_AE and B [Reference Zhang9]. CRF08_BC and CRF01_AE were found to be dominant in IDUs and those with sexually transmitted infections, respectively. After 2006, the main transmission route of HIV-1 in Yunnan changed from intravenous injection to unprotected sexual contact. In the late 2000s, six HIV-1 genotypes were found among recently infected population [Reference Chen10]. Among them, CRF08_BC became the most common genotype, followed by URFs, CRF01_AE, CRF07_BC, subtypes B and C [Reference Chen10]. CRF08_BC was predominant in both heterosexually transmitted populations and IDUs. Strikingly, the frequency of URFs increased dramatically in 12 out of a total of 16 Yunnan's prefectures, suggesting that frequent viral recombination has become a major problem in the growing epidemic.
By the end of 2013, the number of people living with HIV/AIDS (PLWHA) in Yunnan was 79 802, the highest among all provinces in China. Among infected cases diagnosed each year, the prevalence of sexually transmitted infections increased from 63.5% in 2009 to 86.3% in 2013, including an increase from 1.2% in 2009 to 3.7% in 2013 among men who have sex with men (MSM). More cases were reported in central and eastern Yunnan, including the Honghe, Kunming, Wenshan and Qujing prefectures. With the HIV epidemic growing, it is essential to investigate the changing trend of HIV-1 genetics in the province. Meanwhile, antiretroviral therapy (ART) had expanded among PLWHA from 24.3% in 2010 to 53.0% in 2013. With the HIV treatment scaling up, it necessary to conduct a province-wide transmitted drug resistance (TDR) survey to understand the frequency of transmitted drug-resistant viruses. Therefore, we performed a large cross-sectional study of the recently infected population identified in the first quarter of 2014 in the entire Yunnan Province.
Methods
Study participants and sample collection
A total of 2828 HIV/AIDS cases were newly reported between January 2014 and March 2014 in Yunnan Province. The compositions of newly reported HIV/AIDS cases in the first quarter and in the whole year of 2014 showed no differences by means of discovery or infection routes (Supplemental Table S1). The cases with CD4+ T lymphocytes <200 cells/μl or AIDS-defining illnesses were excluded and the remaining 1889 HIV-1-positive cases were subjected to recent HIV-1 infection detection. The adults’ consents were provided by themselves and the juveniles’ consents were provided by their guardians. The study was approved by the Biomedical Ethics Review Committee of Yunnan Province.
Recent HIV-1 infection identified with BED-captured enzyme immunoassay (CEIA)
BED-CEIA was performed according to the manufacturer's instructions (Calypte HIV-1 BED incidence EIA, Calypte Biomedical Corporation, Portland, OR). Test specimens were initially run singly. If the normalised OD (ODn) was >1.2, the specimen was classified as a long-term infection. Specimens with ODn <1.2 were tested again in triplicate to confirm the values. In confirmatory testing, specimens with ODn <0.8 were classified as a recent infection. In China, the window period of BED-CEIA is 168 days [11], which means the recent infection identified by BED-CEIA is within 168 days after seroconversion.
Amplification of HIV-1 gene fragments
Viral RNA was extracted from 140 µl of plasma using the QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. RNA samples were directly subjected to nested polymerase chain reactions (PCR) to generate fragments of gag (HXB2: 781–1861; encoding portions of p17 and p24), pol (HXB2: 2147–3462; encoding the protease and the first 299 residues of reverse transcriptase) and env (HXB2: 7002–7541, encoding the V3–V4 region). The details of amplification and sequencing procedures were previously described [Reference Chen12].
Sequence analysis
The assembly of the different sequences generated from the same gene region of each sample was performed using Sequencher 5.0 (Gene Codes, Ann Arbor, MI). ClustalW multiple alignment and manual editing were performed using Bio-Edit 7.0 software. The reference sequences were obtained from the Los Alamos National Laboratory (LANL) database (http://hiv-web.lanl.gov/content/index), covering the major HIV-1 subtypes/CRFs. Phylogenetic tree analyses were performed using the neighbour-joining method based on the Kimura 2-parameter model with 1000 bootstrap replicates, using MEGA version 6.0 [Reference Tamura13]. To demonstrate possible intersubtype mosaicism, candidate sequences were analysed using the Recombination Identification Programme (RIP, version 3.0; http://hiv-web.lanl.gov).
Bayesian Markov chain Monte Carlo (MCMC) evolutionary analyses
The evolution rate and time of most recent common ancestor (tMRCA) of CRF01_AE, CRF07_BC and CRF08_BC were inferred from gag using Bayesian MCMC method [Reference Ye14]. The sequences of CRF01_AE, CRF07_BC and CRF08_BC identified in the present and previous studies constituted their own datasets for analysis [Reference Chen10, Reference Su15]. Based on the previous study [Reference Chen12], the general time reversible (GTR) model plus a gamma distribution (Γ4) among site rate heterogeneity (I) model (GTR + I + Γ4) was chosen as the nucleotide substitution model. Bayesian MCMC analyses were performed using a Bayesian uncorrelated exponential relaxed molecular clock method in combination with the ‘Bayesian Skyline’ coalescent tree priors under the selected nucleotide substitution model in the BEAST v1.7.4 package [Reference Drummond16]. Each MCMC analysis was run for at least 20 million generations and sampled every 2000 generations. The Maximum Clade Credibility (MCC) tree was obtained by TreeAnnotator v1.7.4 with a burn-in of the initial 10% of generated trees, and examined by FigTree V1.3.1, which was also used to estimate the evolutionary rates and the dates to tMRCA of various nodes on the MCC tree. Combining the parameter log files with the appropriate trees log files, Bayesian skyline plots were constructed with Tracer v1.5.
Geographic distribution analysis of HIV-1 genotypes
HIV-1 genetic geographic distribution was analysed with the public health geographic information system (PHGIS, China CDC). A Dot Density Map was used to display the distribution density of each HIV-1 genotype within Yunnan Province. For each genotype, the number of patients with the genotype in each prefecture was divided by 291 (the number of patients genotyped and included in this study) to obtain the percentage of each genotype in each prefecture. When using PHGIS to map the data, one dot was defined as 0.025% of the population.
Genotypic analysis of HIV-1 drug resistance
The nucleotide sequences of pol gene, containing the full-length protease gene and the first 299 codons of the reverse transcriptase gene, were submitted to Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu). TDR associated mutations were determined with Calibrated Population Resistance (CPR) Tool (Version 6.0). For each transmitted drug-resistant strain, the levels of resistance to commonly used proteases and RT inhibitors were analysed with Genotypic Resistance Interpretation Algorithm.
Sequence data
All the sequences obtained in this study were submitted to GenBank under accession numbers KY244317-KY245056.
Statistical analysis
Statistical analyses were conducted using the SPSS 19.0 statistical analysis software package (SPSS Inc. Chicago, IL). Categorical variables were compared using χ2 tests. All tests were two-tailed and a P value <0.05 was considered statistically significant.
Results
Demographic characteristics of study participants
Among 2828 newly diagnosed HIV-positive samples collected in Yunnan Province during the first quarter of 2014, 347 samples were identified as recent infections by the BED-CEIA assay (within 168 days after seroconversion). The partial gag, pol and env genes of each sample identified as recent infection were amplified and sequenced. Finally, 240 gag, 271 pol and 229 env sequences were obtained. By combining the phylogenetic tree analyses of the three genes, 291 samples were successfully genotyped (Supplementary Figure S1–3). Among the 291 participants, the ratio of males to females was 1.43:1; the mean age was 37.1 years (range: 14–81 years); and 57.4% (167/291) of individuals were of Han ethnicity, while 42.6% of individuals were minority nationalities, including Yi, Hani, Dai, Zhuang, Hui, Lisu, Bai, Wa, Jingpo, Lagu, Miao, Naxi and Deang. Of them, 33.0% (96/291) were single, 53.6% (156/291) were married and 13.4% (39/291) were divorced or widowed. Heterosexual contact was found to be the major transmission route, accounting for 80.8% (235/291), while homosexual contact and intravenous drug injection accounted for 10.0% (29/291) and 7.9% (23/291), respectively (Table 1).
Table 1. Demographic characteristics and HIV-1 subtypes of the study participants
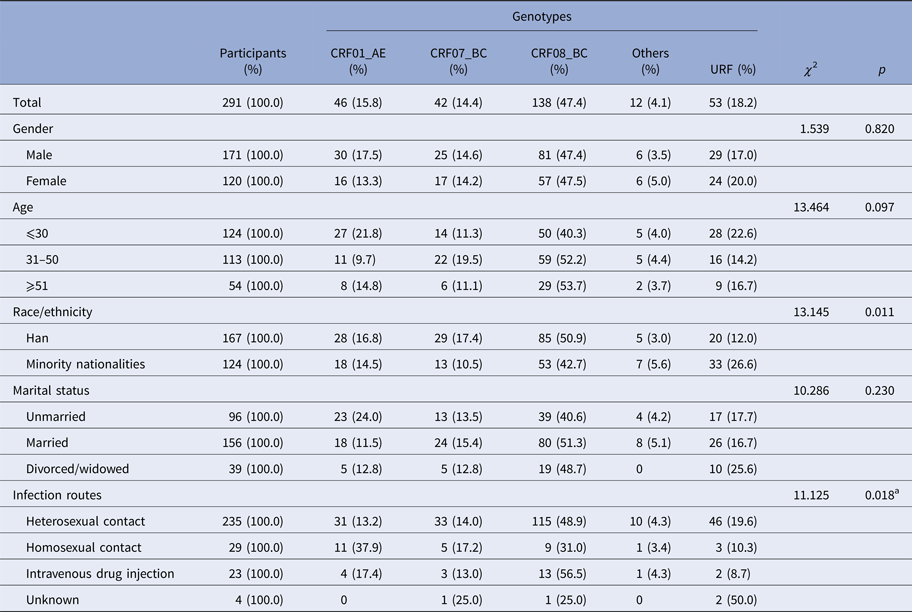
aCompared the constitutions of genotypes between heterosexual and homosexual contacts.
Prevalent HIV-1 genotypes in recently infected population
Based on the sequence data (Supplementary Figure S1–3), two subtypes, five CRFs and five discrete URFs were identified. Among recent infections in this cross-sectional study, CRF08_BC was found to be the most common genotype (47.4%, 138/291), followed by URFs (18.2%, 53/291), CRF01_AE (15.8%, 46/291), CRF07_BC (14.4%, 42/291), subtype C (2.7%, 8/291), CRF55_01B (0.7%, 2/291), subtype B (0.3%, 1/291) and CRF64_BC (0.3%, 1/291). Among the URFs, BC recombinants were the most common recombinant form (77.4%, 41/53), and the other four discrete URFs included BC/CRF01_AE (9.4%, 5/53), CRF07_BC/CRF08_BC (9.4%, 5/53), B/CRF01_AE (1.9%, 1/53) and CRF07_BC/CRF01_AE (1.9%, 1/53).
Demographic distribution characteristic of HIV-1 genotypes
The distribution of HIV-1 genotypes by the participants’ gender, age and marital status showed no statistical differences (Table 1). However, the distribution of HIV-1 genotypes by ethnicity showed a statistical difference (χ 2 = 13.145, P = 0.011). The proportion of URFs in the minority ethnic groups was higher than that in the Han (26.6 vs. 12.0%, χ 2 = 10.235, P = 0.002). Furthermore, the constitutions of HIV-1 genotypes were statistically different between heterosexual transmission and homosexual transmission routes (χ 2 = 20.487, P = 0.011). CRF08_BC and CRF01_AE were the predominant genotypes in heterosexual and homosexual transmission, respectively.
Geographic distribution of HIV-1 genotypes
The geographic distribution of each HIV-1 genotypes was further analysed (Fig. 1). Higher prevalence of CRF08_BC was found in the eastern and central regions, especially in the area centred on Honghe and Kunming. However, CRF08_BC was not identified in the four western prefectures, including Dehong, Nujiang, Diqing and Xishuangbanna. Previously, CRF07_BC was mainly found in the eastern region (Kunming, Qujing and Honghe) [Reference Chen10]. In this study, CRF07_BC was detected in more prefectures (12 prefectures in 2014 vs. four prefectures in 2009). Although CRF01_AE continued to spread throughout the entire province, Kunming was the area with the highest prevalence of this genotype. The distribution of URFs has further extended into 14 prefectures compared with 12 prefectures in 2009 [Reference Chen10]. Significantly, more URFs were identified in the eastern part, including Honghe, Kunming and Wenshan. However, the distribution of subtype B, C and novel CRFs did not show distinct patterns.
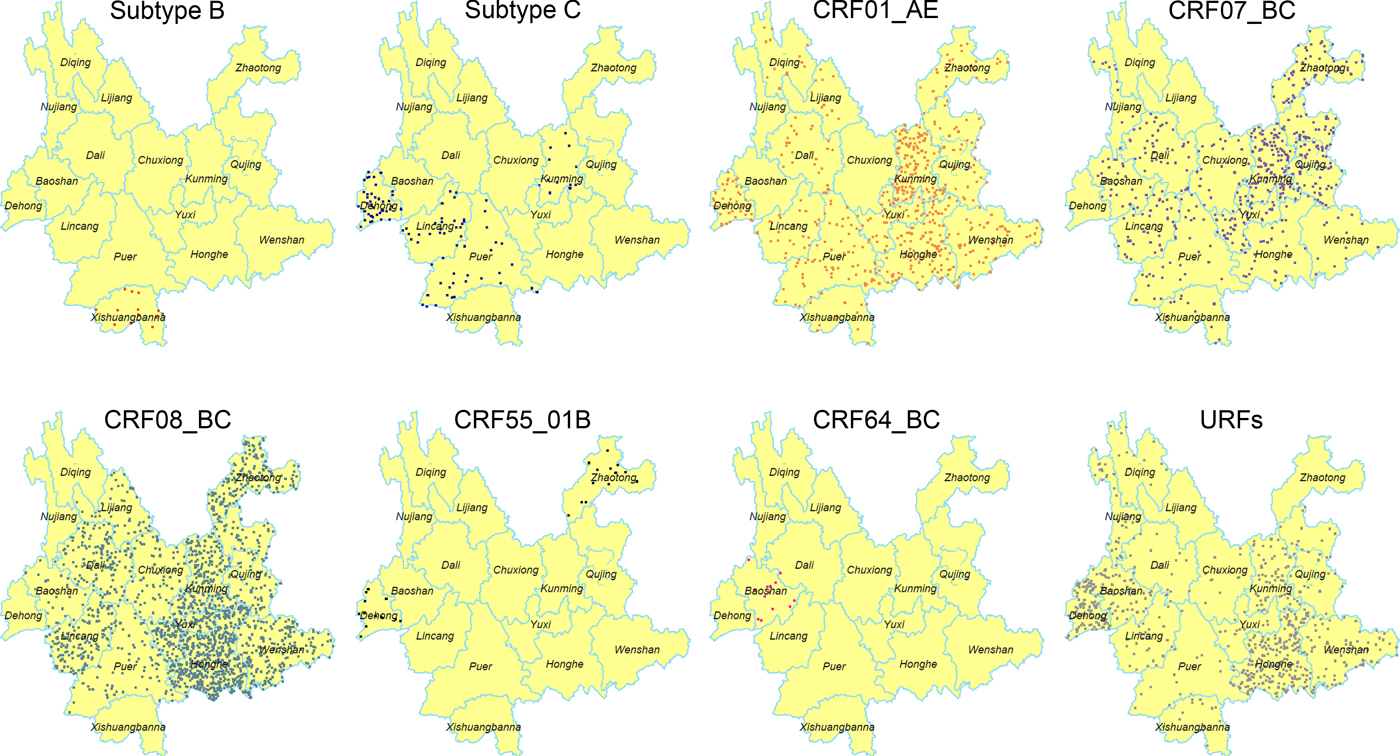
Fig. 1. Geographic distribution of the HIV-1 in Yunnan. Dot Density Map for subtype B, subtype C, CRF01_AE, CRF07_BC, CRF08_BC, CRF55_01B, CRF64_BC and URFs, respectively, which showed the percentage of each genotype in each prefecture. One dot was defined as 0.025% of the population.
It is noteworthy that the distribution of HIV-1 genotypes became more complicated for each prefecture. At least six viral genotypes were found in Kunming, five in the prefectures of Honghe, Puer, Lincang, Dali and Dehong, four in the other seven prefectures (Zhaotong, Qujing, Wenshan, Yuxi, Chuxiong, Xishuangbanna and Baoshan) and two in the three northwest prefectures (Lijiang, Diqing and Nujiang).
Genetic characteristics of the main HIV-1 genotypes
To estimate the evolutionary rates and the time of most recent common ancestor (tMRCA) of the main HIV-1 genotypes circulating in Yunnan, Bayesian MCMC analyses were performed using gag sequences from the present and previous studies[Reference Chen10, Reference Su15]. With the relaxed exponential clock model, the median evolutionary rates of CRF01_AE, CRF07_BC and CRF08_BC were 5.12 × 10−3, 1.14 × 10−2 and 9.87 × 10−3 substitutions site−1 year−1, respectively. Under these substitution rates, the median tMRCA for CRF01_AE, CRF07_BC and CRF08_BC circulating in Yunnan were estimated to be 1983.1, 1992.1 and 1989.5, respectively (Fig. 2).
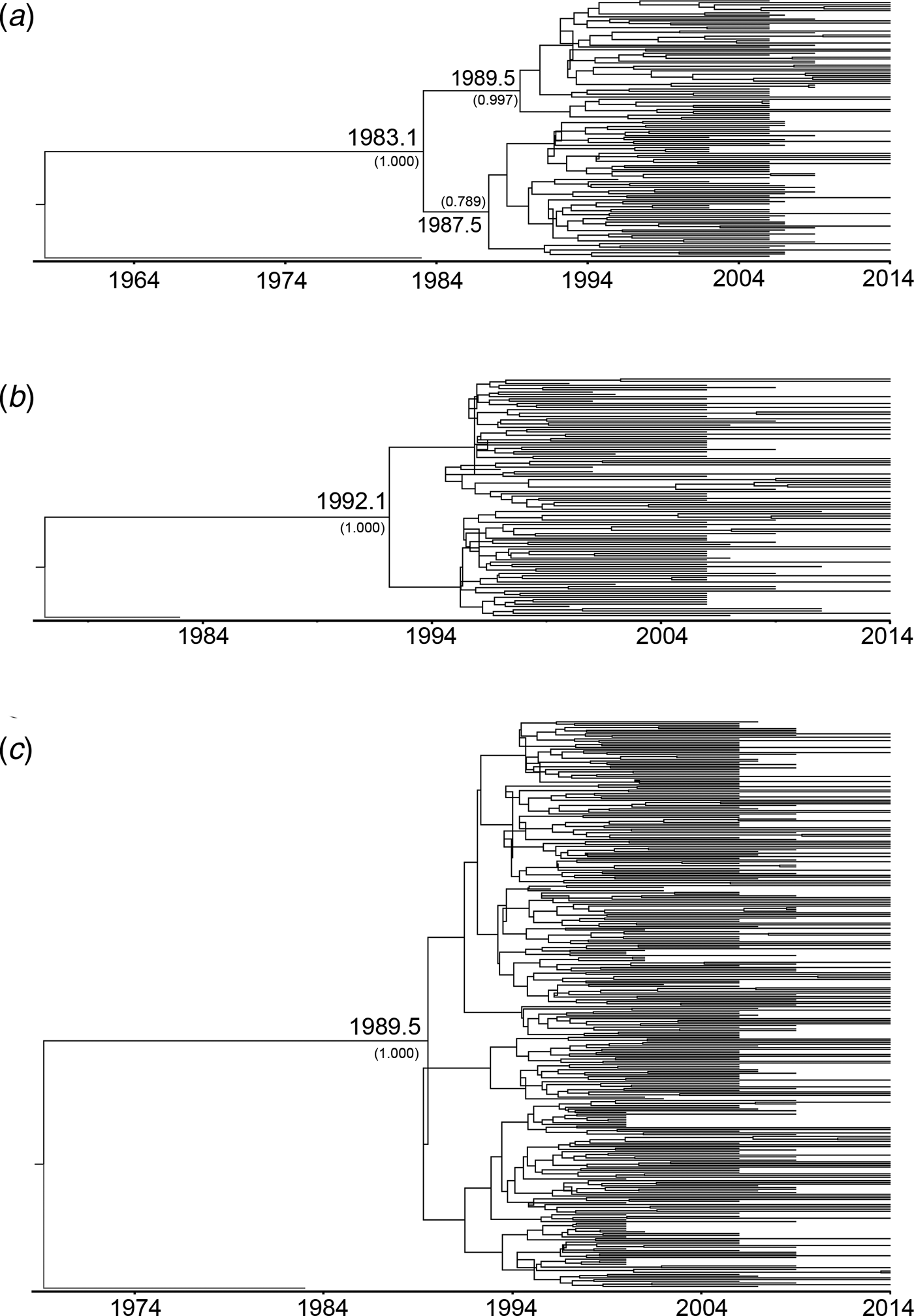
Fig. 2. Maximum clade credibility (MCC) tree representing the rooted genealogy of CRF01_AE, CRF07_BC and CRF08_BC in Yunnan. (A) The MCC tree for CRF01_AE strains. HIV-1 subtype B (B.FR.1983.HXB2-LAI-IIIB-BRU.K03455) was used as the outlier. (B) The MCC tree for CRF07_BC strains. HIV-1 subtype D (D.CD.1983.ELI.K03454) was used as the outlier. (C) The MCC tree for CRF08_BC strains. HIV-1 subtype D (D.CD.1983.ELI.K03454) was used as the outliers. The MCC trees were obtained by Bayesian MCMC analysis based on partial gag gene (HXB2: 2147–3462) implemented in BEAST v 1.7.4. The uncorrelated exponential relaxed molecular clock method was used in combination with the Bayesian Skyline coalescent tree prior under GTR + I + G4 nucleotide substitution model. The branch lengths in the MCC trees reflect the time and the corresponding time-scale is shown at the bottom of the trees. The posterior probabilities of the key nodes and the tMRCA medians for the interested nodes are indicated.
Demographic history of the major HIV-1 genotypes
To further investigate the demographic history of CRF01_AE, CRF07_BC and CRF08_BC in Yunnan, we analysed the changes in effective population size (EPS) (the number of infected individuals contributing to HIV transmission) by Bayesian Skyline Plots. For CRF01_AE (Fig. 3a), the initial growth phase began in the late 1980s, followed by an exponential growth from 1991 to 1999 (red area). The EPS reached its peak around 2004, followed by a slight decline from 2005 to 2008 (green area) and became plateaued from 2009 onwards. For CRF07_BC (Fig. 3b), the EPS initially increased in the early 1990s, then underwent an exponential increase around 1994–1999 and reached a stationary phase thereafter. For CRF08_BC (Fig. 3c), the EPS initially increased in the early 1990s, then underwent two exponential growth phases from 1994 to 1998 and 2001 to 2002, and became constant from 2004 onwards. It appears that the second exponential growth resulted in CRF08_BC becoming the predominant lineage in Yunnan.
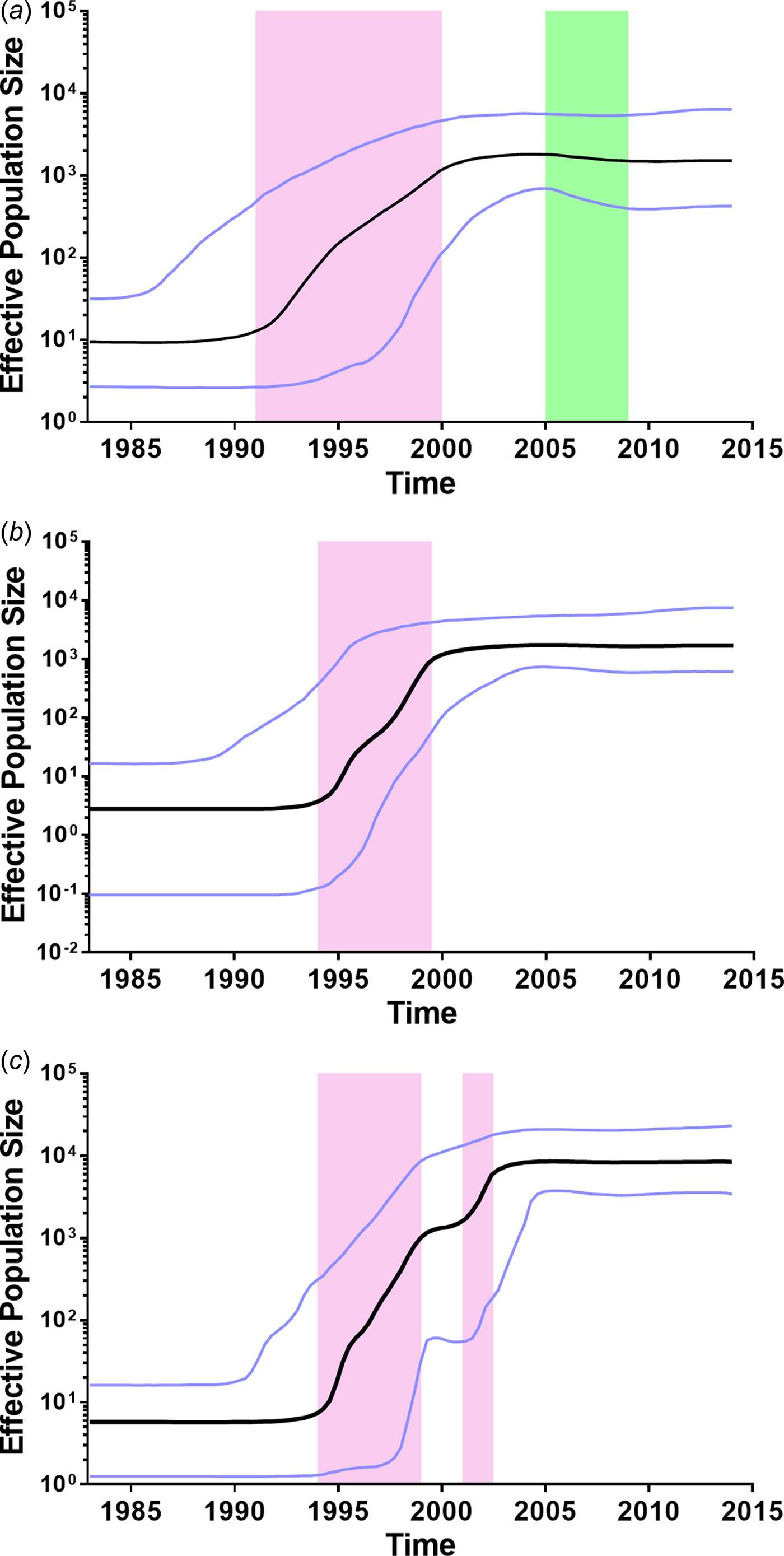
Fig. 3. Bayesian skyline plots of effective population sizes (EPSs) for the main HIV-1 strains in Yunnan. (A) Bayesian skyline plot of EPS for CRF01_AE. (B) Bayesian skyline plot of effective population size for CRF07_BC. (C) Bayesian skyline plot of effective population size for CRF08_BC. The vertical ruler on the left (Y axis) measures the EPS while the horizontal scale on the bottom measures time from the present (right) to the past (left). The black line represents the median of EPS through time. The purple lines represent the lower and higher 95% highest posterior density. The pink area represents that EPS exponentially increases. The green area represents that EPS declines.
Genotypic analysis of TDR
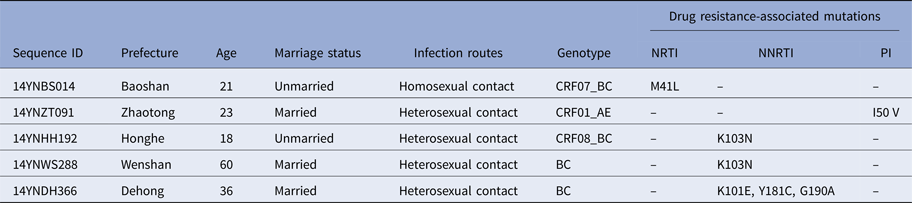
To determine whether TDR-associated HIV-1 mutants were rising in recent years, we analysed the 271 pol sequences obtained. Results in Table 2 show that five participants (1.8%) harboured viral mutants with TDR-associated mutations. The proportions of sequences with resistance to nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors (PIs) were 0.4% (1/271), 1.1% (3/271) and 0.4% (1/271), respectively. Among the five participants with TDR-associated mutations, one (14YNDH366) had three drug-resistant mutations (K101E, Y181C and G190A) while the other four had single mutations.
Table 2. Demographic characteristics of five individuals harbouring transmitted drug resistance-associated mutations
Discussion
In the present study, we conducted a large-scale, province-wide HIV-1 genetic study in recently infected population in Yunnan which is most severely affected by HIV/AIDS in China. Based on the cumulative HIV-1 sequences data, we further investigated the genetic characteristics and demographic history of the main HIV-1 strains circulating in Yunnan. In addition, the analysis of HIV-1 TDR in this province was conducted for the first time.
The ranking of the predominant HIV-1 genotypes in Yunnan was found in the following order: CRF08_BC > URFs > CRF01_AE > CRF07_BC. Compared with the data collected in 2009, the proportions of CRF08_BC, CRF01_AE and CRF07_BC showed no statistical difference (CRF08_BC: 47.4% in 2014 vs. 40.8% in 2009, χ 2 = 1.605, P = 0.244; CRF01_AE: 15.8% in 2014 vs. 18.5% in 2009, χ 2 = 0.457, P = 0.571; CRF07_BC: 14.4% in 2014 vs. 9.2% in 2009, χ 2 = 2.175, P = 0.0.158;) [Reference Chen10], which suggested that these three lineages remained relatively stable. However, the proportion of URFs decreased significantly (18.2% in 2014 vs. 27.7% in 2009, χ 2 = 4.843, P = 0.029) probably due to the reason that some of the previous URFs were identified as novel CRFs. In the last five years, several novel CRFs were identified from western Yunnan, including CRF57_BC [Reference Li17, Reference Wei18], CRF62_BC [Reference Wei19], CRF64_BC [Reference Hsi20] and CRF65_cpx [Reference Feng21]. CRF55_01B was originally identified from MSM in South and Central China [Reference Han22], but here, this genotype was identified in both homosexual and heterosexual individuals, suggesting that this CRF is no longer limited to MSM and has spread through the unprotected heterosexual route.
URFs contribute to the increased diversity of HIV-1 genetics. Among URFs found in Yunnan, URFs_BC was the most prevalent recombinant form, accounting for ~70% of total URFs. A recent phylogenetic study showed that URFs_BC had already emerged at the early stage of the HIV-1 epidemic in Yunnan (in the late 1980s), from which CRF07_BC and CRF08_BC arose in the early 1990s [Reference Feng7]. Furthermore, CRF57_BC and CRF64_BC appear to have emerged in the early 1990s, while CRF62_BC was estimated to have originated in the early 2000s [Reference Feng7]. In present, the proportion of URFs_BC remains high in Yunnan, which may suggest that some novel CRFs_BCs have yet to be identified.
In the late 2000s, heterosexual contact and intravenous drug use were the dominant modes of HIV transmission among recently infected people [Reference Chen10]. At that time, CRF08_BC was the predominant viral genotype in both heterosexuals and IDUs. In recent years, a dramatic increase in the HIV-1 epidemic was evident amongst MSM. As a result, homosexual contact overtook intravenous drug use in terms of HIV-1 transmission. We found in this study that CRF08_BC and CRF01_AE were the predominant genotypes in heterosexual and homosexual contact, respectively. Moreover, the constitutions of HIV-1 genotypes were statistically different between these two populations. These suggested that unique transmission characteristics exist among MSM, the characteristics of HIV molecular epidemics among MSM need to be further studied.
Traditionally, CRF07_BC and CRF08_BC were highly prevalent in eastern Yunnan, but CRF08_BC was distributed more widely than CRF07_BC. CRF01_AE was first found in the western prefectures then spread to the eastern prefectures [Reference Zhang9]. Compared with our study in 2009, CRF07_BC and CRF01_AE have further spread. However, they appear to spread in the opposite directions, where CRF07_BC went to the west, and CRF01_AE moved east. Similarly, the distribution of URFs has also enlarged and can be found in 14 prefectures. As a result, CRF08_BC, URFs, CRF01_AE and CRF07_BC were widely distributed in Yunnan. However, the areas with higher prevalence of CRF08_BC, URFs, CRF01_AE and CRF07_BC were in the central and eastern regions of Yunnan, such as Honghe, Kunming, Wenshan and Qujing, where more recent infections were reported.
Further, we retrospectively analysed the history of CRF01_AE, CRF07_BC and CRF08_BC in Yunnan. CRF01_AE was considered to be introduced from Thailand [Reference Cheng6, Reference Feng23]. Here, the estimated tMRCA of CRF01_AE sequences in Yunnan was 1983.1, which is similar to that reported in Thailand [Reference Perez-Losada24], suggesting that CRF01_AE in Yunnan and Thailand could share the common ancestor. In Yunnan, CRF01_AE was first reported among FSWs who had returned from Thailand to Yunnan [Reference Feng7]. After that, CRF01_AE became the predominant strain in FSWs in the early stage of the HIV-1 epidemic [Reference Lu1, Reference Zhang9]. As a bridging population, FSWs introduced this strain into heterosexually-infected individuals. As a result, in the early 2000s, the proportion of CRF01_AE among all subtypes/CRFs reached 40.5%, and 85.4% of CRF01_AE infection were acquired through heterosexual transmission [Reference Zhang9]. These data support that the EPS of CRF01_AE increased exponentially during 1991–1999, and reached the steady state in the early 2000s. However, a slight decline of EPS appeared between 2005 and 2008. The surveillance data showed that HIV incidence significantly decreased in FSWs for 2007–2010 (from 0.71 to 0.31%) [Reference Yang25], probably due to health education and condom promotion in the high-risk populations. These might explain the slight decline of EPS during in this period of time. In the recent years, the annual rate of newly reported HIV cases attributed to homosexual contact has increased (from 1.2% in 2009 to 4.3% in 2014), however, which was still far lower than that attributed to heterosexual contact (from 62.3% in 2009 to 85.2% in 2014). Although CRF01_AE was the predominant strain among MSM, the proportion of CRF01_AE infections acquired through homosexual contact was relatively low among all CRF01_AE infections.
CRF07_BC and CRF08_BC were speculated to originate in Yunnan Province [Reference Feng7, Reference Tee8]. In this study, the estimated tMRCA for CRF07_BC and CRF08_BC were approximately 1992.1 and 1989.5, respectively, which were a little earlier than their counterparts reported previously [Reference Feng7, Reference Tee8]. Both CRF07_BC and CRF08_BC underwent an exponential growth in the late 1990s. During the exponential growth period, the average growth rates of EPS for CRF07_BC and CRF08_BC (0.420 and 0.414 log/year) were faster than that for CRF01_AE (0.217 log/year). However, CRF08_BC underwent a second exponential growth from 2001 to 2002, which made the EPS of CRF08_BC exceed those of CRF01_AE and CRF07_BC. Perhaps, in the early 2000s, the bridging population drove the influx of CRF08_BC from IDUs into the heterosexually transmitted population, such as IDUs who visit FSWs and FSWs who inject drugs.
Based on the recommendations by World Health Organization [Reference Bennett26], we have performed HIV-1 drug resistance (HIVDR) threshold surveys in some key regions affected by HIV in Yunnan, including Dehong, Kunming, Honghe, Wenshan and Dali since 2009. The results showed that the overall prevalence of transmitted HIV-1 drug resistance kept a low level (<5%). To minimise the use of resources, the HIVDR threshold survey method takes the newly diagnosed HIV-infected individuals aged <25 years as recent infections. Our study used the BED-CEIA to screen samples for province-wide TDR survey, by which the selection bias could be properly controlled. Our results, therefore, confirmed the low prevalence of transmitted HIVDR in Yunnan, which guaranteed the effectiveness of the regimens of Chinese national free ART programme. To control the transmitted HIVDR, the priority is to improve the patient management.
In this study, we tracked the changes of HIV-1 genetic characteristics by performing consecutive cross-sectional HIV-1 molecular epidemiological survey among the recently infected population. CRF08_BC, URFs, CRF01_AE and CRF07_BC were still the predominant strains, and their coverage further enlarged. Meanwhile, some novel CRFs were found in recent infections, which contribute to the HIV-1 genetic diversity in Yunnan. Demographic history of the main HIV-1 strains suggested that CRF01_AE, CRF07_BC and CRF08_BC underwent a rapid-growth in the 1990s. To date, the EPSs of CRF01_AE, CRF07_BC and CRF08_BC remained stable at a high level. The prevalence of transmitted HIV-1 drug resistance kept a low level in Yunnan. These findings provide valuable information to develop effective measures to prevent new infections.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268818000389.
Acknowledgements
We are very thankful to the staff at the local Centers for Disease Control and Prevention for their assistance in coordinating samples collection.
Financial support
This work was supported by the National Natural Science Foundation of China (grant no. 81560327), the research project for Yunnan science and technology plan (grant no. 2015FB200), Yunnan health science and technology plan project (grant no. 2014NS348) and the Major Project of China's ‘Twelfth Five-Year Plan’ for Science and Technology Development (2013ZX10004-906).
Conflict of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.








