INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading nosocomial pathogen that was first reported in Canada in 1981 [Reference Low1] and has since disseminated nationwide [Reference Simor2, Reference Simor3]. Over the last decade, community-associated MRSA (CA-MRSA) infections have rapidly emerged on a global scale in many aboriginal reserves, the military, intravenous drug users, the homeless, penitentiaries, amateur and professional sports teams, day-care centres, and schools [Reference Aiello4–Reference Ofner-Agostini11]. Of great concern is that these CA-MRSA strains are: (1) causing infections in often young otherwise healthy individuals with no predisposing nosocomial risk factors [Reference Mulvey10], (2) associated with more severe disease [Reference Adam, McGeer and Simor12–Reference Vayalumkal14], and (3) now entering and disseminating within hospitals [Reference Seybold15, Reference D'Agata16].
In the 1990s, CA-MRSA had been reported only sporadically in Canada [Reference Embil17, Reference Shahin18]. However, since that time two CA-MRSA strains have emerged in Canada, CMRSA7 (USA400) and CMRSA10 (USA300) [Reference Christianson19]. CMRSA10 has emerged predominantly in the western provinces of Canada and high incidences have been reported in the homeless, incarcerated, and intravenous drug user populations in the Calgary [Reference Gilbert7] and Vancouver [Reference Al-Rawahi5, Reference Al-Rawahi20] health regions. In comparison to the incidence of CA-MRSA infections in the large urban centres across Canada, which is represented through the ongoing efforts of the Canadian Nosocomial Infection Surveillance Program [Reference Simor2, Reference Simor3], very little attention has been directed at the emerging problem of CA-MRSA in northern communities in Canada. CMRSA7 was first reported in Manitoba as an outbreak in the southern portion of that province in the late 1990s, but has since spread to the northern regions of the province between 2000 to 2004 [Reference Kurbis and Wylie21, Reference Wylie and Nowicki22]. The emergence of CMRSA7 was thereafter seen in a central eastern Saskatchewan community adjacent to the Manitoba border [Reference Mulvey10] and has since disseminated into all three northern health regions of Saskatchewan [Reference Irvine23] as well as Nunavut [Reference Dalloo24]. The emergence of CMRSA7 in many of these northern communities is causing primarily skin and soft tissue infections in children with crude rates of infection as high as 196 cases/10 000 in some northern Saskatchewan health regions [Reference Irvine23].
The factors influencing the emergence and spread of CA-MRSA may be different in these northern communities compared to factors identified in studies involving large urban centres where cultural, socioeconomic, medical, and environmental conditions are different. To better understand the dissemination of CA-MRSA in remote northern communities a case-control study was undertaken to identify potential risk factors associated with the acquisition and dissemination of CA-MRSA in two selected northern Saskatchewan health regions.
METHODS
Setting
The study was a prospective cohort of CA-MRSA and community-associated methicillin-susceptible S. aureus (CA-MSSA) infections identified from September 2004 to March 2007 within selected communities of the Kelsey Trail Health Region (KTHR) and the Mamawetan Churchill River Health Region (MCRHR). The KTHR encompasses a large geographic area of east-central Saskatchewan, ~44 369 km2, and has an estimated population of 42 098 (0·9 per km). The MCRHR is located in the north-eastern area of Saskatchewan and encompasses the largest health region in the province, ~93 427 km2, and has an estimated population of 22 427 (0·2 per km) (www.health.gov.sk.ca/covered-population2008).
Identification of MRSA and MSSA isolates
Isolates were identified as S. aureus using standard microbiological methodologies at the Nipawin Hospital Laboratory (NHL), Sandy Bay Health Center (SBHC), and the Saskatchewan Disease Control Laboratory (SDCL). MRSA was defined as isolates of S. aureus showing growth on oxacillin agar screen plates (Mueller–Hinton agar supplemented with 4% NaCl and oxacillin, 6 μg/ml) incubated at 35°C for 24 h. All isolates identified as phenotypically positive for MRSA from the NHL or SBHC were sent to SDCL for confirmation by detecting for the presence of mecA and nuc genes using PCR [Reference McDonald25].
Case definitions
In this study, cases (CA-MRSA) and controls (CA-MSSA) were defined as a laboratory-confirmed community-acquired infection according to the Center for Disease Control and Prevention (CDC) definitions (non-hospitalization >48 h before the positive MRSA culture; no dialysis, surgery, residence in a long-term care facility, or hospitalization in the past year, no prior MRSA infection in the past year, and no percutaneous device or indwelling catheter) [Reference Naimi26]. Individuals identified as potential study participants were contacted by community healthcare workers directly to determine their interest in participating in the study. Patients consenting to participate were interviewed using a standard investigation form (available at www.narp.ca), which captured demographic and exposure to possible risk factors. The study was approved by the research ethics boards of the University of Manitoba, the University of Saskatchewan, and Health Canada.
Isolate characterization
All CA-MRSA and CA-MSSA isolates were submitted to the National Microbiology Laboratory (NML) for additional molecular characterization, which included pulsed-field gel electrophoresis (PFGE) and spa-typing performed as previously described [Reference Mulvey27, Reference Harmsen28]. PCR for detection of the mupA gene was as previously described [Reference Anthony29]. Antimicrobial susceptibility of isolates was determined using standard broth microdilution panels according to CLSI guidelines [30]. Breakpoints used for fusidic acid and mupirocin resistance, which were not provided in the CLSI guideline, were as previously described [Reference Skov, Frimodt-Moller and Espersen31, Reference Walker32]. Inducible resistance to clindamycin in macrolide-resistant strains of CA-MRSA and CA-MSSA was detected by a standardized disk approximation test [30].
Statistical analysis
Data was entered into Epi-Info version 3.4.3 (CDC, USA). Univariate analysis was performed to compare the demographic variables, site of infection, and medical history between the CA-MRSA and CA-MSSA cases. Fisher's exact test or χ2 tests where appropriate were used to compare categorical variables. Unpaired Student's t test was used for continuous data. Multivariate logistic regression analysis was used to determine characteristics associated with acquisition of CA-MRSA or CA-MSSA. Stepwise logistic regression models for P<0·25 were used in the multivariate analysis. All tests were two-tailed, and a P value of <0·05 was considered statistically significant. Multivariate analysis was performed using SPSS version 11.0 (SPSS Inc., USA).
RESULTS
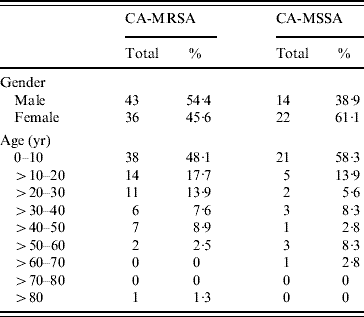
A case-control study was undertaken comparing patients who had acquired either a CA-MRSA (case) or CA-MSSA (control) infection in order to identify potential risk factors for acquiring CA-MRSA infections. A total of 115 patients, representing 79 CA-MRSA and 36 CA-MSSA infections, fulfilled the screening criteria and provided consent to participate in this study. No significant differences were observed when comparing gender or age of patients who had acquired either a CA-MRSA or CA-MSSA infection (Table 1). The majority of CA-MRSA (79·7%) and CA-MSSA (77·8%) infections were in individuals aged <30 years (Table 1). Skin and soft tissue was the predominant site of infection for both CA-MRSA (67·1%) and CA-MSSA (55·6%), but no significant differences were observed when comparing specific anatomical sites of infection (data not shown). Of note, only CA-MRSA isolates (7·7%, P=0·10) were obtained from the buttock region.
Table 1. Gender and age distribution of CA-MRSA and CA-MSSA infections
The investigation surveys used to identify potential risk factors for the acquisition of CA-MRSA did not reveal any statistical differences between a CA-MRSA and CA-MSSA infection (Table 2). For individuals with CA-MRSA infections, slightly higher proportions who were smokers (OR 2·12, 95% CI 0·75–6·14, P=0·12) and lower proportions with access to indoor plumbing (OR 0·3, 95% CI 0·06–1·23, P=0·06) were noted. Shared common risk factors included existing skin conditions, exposure to someone with a skin condition or MRSA infection, scratches and insect bites, exposure to healthcare workers, previous antibiotic usage, and overcrowding (Table 2). In regard to antimicrobial usage, the average number of antimicrobials prescribed in the previous year was 2·5 (median 2, range 0–10) and 1·7 (median 2, range 0–4) for patients with either a CA-MRSA or CA-MSSA infection, respectively. Concerning overcrowding, the average number of people per household was 6·3 (median 7, range 1–14) and 5·4 (median 5·5, range 1–9) for CA-MRSA and CA-MSSA infections, respectively.
Table 2. Investigative forms to identify potential risk factors for CA-MRSA infections (percentage in parentheses calculated based on total Yes/No respondents)

COPD, Chronic obstructive pulmonary disease; CHF, congestive heart failure.
A high proportion of the patients, 88·7% for CA-MRSA and 79·3% for CA-MSSA, were prescribed antimicrobials for their infection. The descending order of the top three classes of antimicrobials empirically prescribed alone or in combination with another drug for the treatment of CA-MSSA infections was a folate pathway inhibitor [trimethoprim–sulfamethoxazole (TMP–SXT)] (50%)>β-lactam (including β-lactamase-sensitive and -resistant penicillins and/or cephalosporins) (33·3%)>macrolide (25%). The descending order of the top three empirically prescribed antimicrobial classes alone or in combination with another drug for the treatment of CA-MRSA infections in the region were β-lactam (including β-lactamase-sensitive and -resistant penicillins and/or cephalosporins) (57·1%)>TMP–SXT (28·6%)>mupirocin (14·3%)⩾macrolide (14·3%).
CA-MRSA and CA-MSSA isolates were characterized by PFGE and spa-typing. For CA-MRSA, Canadian MRSA PFGE epidemic type CMRSA7 (USA400/MW2) was the predominant clone in these regions, accounting for 74·7% of all tested CA-MRSA isolates. Of the 59 CMRSA7 isolates, three spa types were identified (t128, n=48; t127, n=7; and t1788, n=4). The remaining two CA-MRSA PFGE clusters, CMRSA2 (USA100/800) and CMRSA10 (USA300), represented 22·8% and 2·5% of the remaining isolates, respectively (Fig. 1). The spa types for the CMRSA2 and CMRSA10 isolates were t311 and t008, respectively. Of the CA-MRSA isolates, 46·8% were found to harbour the genes encoding for PVL, which were only identified amongst the CMRSA7 (35/59) and CMRSA10 (2/2) isolates. The 24 CMRSA7 PVL-negative isolates were identified amongst various PFGE fingerprint patterns, but were indistinguishable from the PVL-positive isolates of the same PFGE fingerprint pattern.

Fig. 1. Pulsed-field gel electrophoresis (PFGE) dendrogram of the CA-MRSA (n=79) and CA-MSSA (n=36) isolates. MRSA and MSSA PFGE clusters are indicated by the shaded blocks defined by >80% similarity index and spa type. Canadian epidemic MRSA PFGE types are indicated in text. PFGE clusters of MSSA isolates were arbitrarily labelled 1–5 for this study.
In comparison to CA-MRSA, CA-MSSA were genetically more diverse with a number of different PFGE strain types identified. Eight CA-MSSA isolates displayed unique PFGE fingerprints, whereas the fingerprints for the remaining 28 isolates were distributed in five PFGE clusters (MSSA1-5) (Fig. 1). MSSA4 was the predominant CA-MSSA strain in the region, accounting for 25% of the typed CA-MSSA isolates, and were all found to be t159 by spa-typing. A second cluster, MSSA1, accounted for 19·4% of the CA-MSSA isolates and the predominant spa type amongst these isolates was t084. Two of the remaining CA-MSSA PFGE fingerprint clusters, MSSA2 (spa types t311, t002) and MSSA5 (spa types t012, t018) were related to Canadian PFGE MRSA epidemic types CMRSA2 and CMRSA4, respectively, by both PFGE and spa-typing [Reference Golding33]. All CA-MSSA isolates were negative for the genes encoding PVL using PCR.
Of the 11 antimicrobials tested, no resistance to vancomycin, linezolid, TMP–SXT, and tetracycline was observed for the CA-MRSA and CA-MSSA isolates (Table 3). CA-MRSA were statistically more likely to be resistant to ciprofloxacin (OR 4·32, 95% CI 0·87–29·0, P=0·04) and mupirocin (OR 13·92, 95% CI 4·5–45·0, P<0·001). All mupirocin-resistant CA-MRSA isolates displayed high level resistance (⩾256 μg/ml) in comparison to only 36% (5/14) of the CA-MSSA isolates. Select high level mupirocin-resistant isolates were all found to harbour the mupA gene by PCR. The minimum inhibitory concentration for low-level mupirocin-resistant CA-MSSA isolates ranged from 4–16 μg/ml and all were negative by PCR for the mupA gene. In addition to mupirocin, resistance to another topical antimicrobial, fusidic acid, was relatively high in both CA-MRSA (21·5%) and CA-MSSA (19·4%) isolates from these regions. High levels of resistance to erythromycin and clindamycin was also noted for both CA-MRSA (30·4%/22·8%) and CA-MSSA (41·7%/27·8%) isolates. Inducible resistance to clindamycin accounted for 12/18 (66·7%) and 7/11 (63·6%) of the clindamycin-resistant CA-MRSA and CA-MSSA isolates, respectively.
Table 3. Antimicrobial susceptibilities for CA-MRSA and CA-MSSA infections
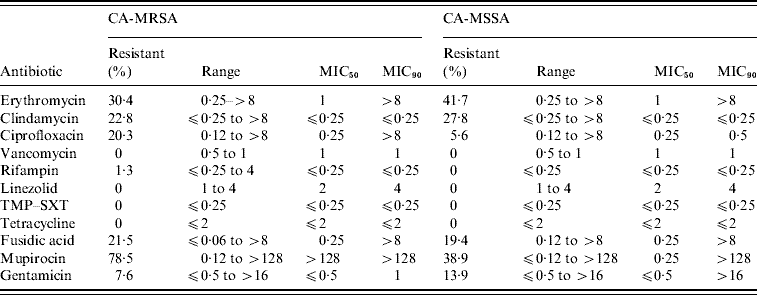
TMP–SXT, Trimethoprim–sulfamethoxazole; MIC, minimum inhibitory concentration.
DISCUSSION
Anecdotal evidence from northern Saskatchewan and other Canadian communities suggests that physicians and prescribing nurses frequently opt to empirically prescribe antimicrobials without obtaining laboratory identification of the infecting organism or drug resistance. It has therefore been speculated that liberal antimicrobial prescribing practices in northern communities may contribute to community acquisition of CA-MRSA. In this study, exposure to antibiotics in the year prior to acquiring the infection was very high in people with either a CA-MRSA (88·6%) or CA-MSSA (97·2%) infection (Table 2). Data obtained for the treatment of CA-MRSA or CA-MSSA infections revealed that a high proportion of individuals were prescribed antimicrobials. Due to the high usage of β-lactams, appropriate antibiotics were more likely to be given to patients with a MSSA infection, which could be contributing to treatment failures, recurrent infections, and/or further dissemination of CA-MRSA within these communities.
It should be noted that this study had some limitations, including the small number of CA-MSSA infections (n=36) that reduced the study's statistical power and also prevented the use of matching criteria. In addition, the average follow-up time was 6·1 months from time of positive culture to completed questionnaires, which could present some recall bias into the study. Unique risk factors for the acquisition of a CA-MRSA infection vs. a CA-MSSA infection were not identified, which coincides with a recently reported community-onset MRSA infection Danish study [Reference Bocher34]. In the Danish study, the only two significant risk factors identified included non-Danish origin and prior hospitalization for more than 7 days within 6 months before infection, the latter of which would have been screened out in our study. Common risk factors for CA-MRSA and CA-MSSA identified here, including younger age groups, exposure to healthcare workers, previous antibiotic usage, chronic skin conditions, and household contact to person with MRSA and exposure to person with a skin condition (Table 3), were similar to a recent investigation of CA-MRSA in a remote northern community in Nunavut, Canada [Reference Dalloo24]. As with this study, the predominant CA-MRSA clone identified in the Nunavut community was CMRSA7 (USA400) [Reference Dalloo24].
Community-associated outbreaks in Canada's larger urban centres have predominantly involved CMRSA10 (USA300). The most common risk factors reported for CMRSA10 infections have included history of illicit drug use, homelessness, and incarceration [Reference Al-Rawahi5, Reference Gilbert7], which was not pertinent in this study nor in the Nunavut study [Reference Dalloo24] as risk factors for CMRSA7 infection. These different risk factors might be contributing to the separate dissemination of CMRSA7 throughout the northern prairies and Nunavut, whereas CMRSA10 is predominating in western Canada's larger urban centres. A second possibility, which remains to be examined, is that differences in host genetics, immunity, and/or microbiomes could result in enhanced susceptibility of a population to a particular CA-MRSA strain type.
Similar risk factors, identified for both CA-MRSA and CA-MSSA infections in our study, and recently in Denmark [Reference Bocher34], suggest that standard hygienic measures and proper treatment guidelines are likely to be beneficial in controlling both CA-MRSA and CA-MSSA. Similar strategies have previously been successful in limiting CA-MRSA transmission in prison inmates [Reference Main9] and sports teams [Reference Romano, Lu and Holtom35]. To address this issue in northern Saskatchewan, treatment algorithms and many educational materials, such as hand-washing posters, patient pamphlets, community presentations, radio advertisements, podcasts, and newly developed educational tools for school-aged children (Germs Away and a computer flash animation game) have been provided throughout many northern communities and schools in Saskatchewan. These materials are all freely available (www.narp.ca) and will hopefully aid in increasing awareness of the importance of proper antimicrobial usage and hygiene in diminishing the spread of disease.
ACKNOWLEDGEMENTS
Funding was provided by the Canadian Institutes of Health Research and the Public Health Agency of Canada.
DECLARATION OF INTEREST
None.





