INTRODUCTION
Noroviruses (NoVs) represent one of the most common causes of enteric disease in humans [Reference Koopmans1, Reference Zeng2]. The prevalence of NoV in children with acute gastroenteritis ranges from 6% to 48% [Reference Koopmans1–Reference Ferreira3]. NoVs cause significant mortality and morbidity worldwide with 200 000 deaths each year in children aged <5 years, 64 000 paediatric hospital admissions and 900 000 paediatric hospital visits in developed countries each year [Reference Patel4].
NoVs, family Caliciviridae, are non-enveloped viruses with a single-stranded, positive-sense, polyadenylated RNA genome of about 7500 nucleotides in length [Reference Green, Knipe and Howley5], organized into three open reading frames (ORFs), ORF1, 2, and 3, which encode a polyprotein including the RNA-dependent RNA polymerase (RdRp), the major capsid protein, and a small capsid protein, respectively.
NoVs encompass five distinct genogroups (GI–GV) with GI, GII and GIV infecting humans. The NoV genogroups are further divided into genotypes and variants based on sequence diversity [Reference Green, Knipe and Howley5]. Along with point mutations, recombination events seem to contribute largely to the genetic variability of NoVs [Reference Bull6]. The ORF1/ORF2 junction region is a recombination hotspot and this creates various combinations of RdRp and capsid types [Reference Bull6]. Multi-target characterization of NoVs in the RdRp region (diagnostic regions A and B) [Reference Ando7–Reference Yuen10] and capsid gene (diagnostic regions C–D) [Reference Vinjé, Hamidjaja and Sobsey11–Reference Kojima14] is therefore necessary to characterize precisely NoVs.
Thus far, 14 GI and 29 GII RdRp genotypes, and eight GI and 23 GII capsid genotypes have been described. RdRp genotypes are designated by a capital P (for ‘polymerase’), followed by an arabic number, i.e. GII.P4, in order to avoid confusion with capsid type nomenclature [Reference Kroneman15].
Over the last decade the most common combinations of RpRd/capsid genotypes in the circulating NoV strains were GII.P4_GII.4 and GII.Pb_GII.3 [Reference Hoa16]. GII.4 NoVs are highly heterogeneous genetically and evolve quickly, generating new variants every 2 or 3 years [Reference Siebenga17–Reference Bull20]. Some GII.4 variants (Farmington Hills 2002, Hunter 2004, Yerseke 2006a, Den Haag 2006b, Apeldoorn 2008, New Orleans 2009, Sydney 2012) in the last decade have reached a global distribution [Reference Hoa16, Reference van Beek21].
While reports on NoV infections/outbreaks are numerous, studies describing the molecular epidemiology of NoVs associated with sporadic gastroenteritis are scanty, usually spanning a short time frame [Reference Hoa16]. However, gathering information on the epidemiology of NoVs is pivotal for tracking the origin of infections, understanding the dynamics of NoV circulation and devising appropriate public health intervention strategies. Patients hospitalized with gastroenteritis are a good target for gathering epidemiological information on enteric pathogens circulating in the community.
This study describes the results of a 5-year (2007–2011) hospital-based surveillance activity for sporadic NoV infections associated with gastroenteritis in the area of Parma, Northern Italy. Molecular characterization in the ORF1 and ORF2 of a representative selection of NoV strains was achieved in order to generate a reliable picture of NoV molecular epidemiology.
MATERIALS AND METHODS
Stool specimens
Stool samples of 2834 children (age range 0–169 months, median age 33 months) with acute gastroenteritis attending the Maternal Infantile Department of the University Hospital of Parma, Northern Italy, as inpatients (84·7%) and outpatients (15·3%) during January 2007–December 2011 were submitted for virological investigations at the Virological Unit of the Department of Pathology and Laboratory Medicine of the same University Hospital.
The faecal specimens were clarified in 10% (w/w) suspensions in phosphate-buffered saline (PBS, pH 7·2) and analysed by electron microscopy (EM), latex agglutination for group A rotavirus (RV) and adenovirus (AdV) (Orion Diagnostica, Finland), and inoculated onto cell cultures as previously reported [Reference Medici22]. Cytopathogenic agents were identified by EM, neutralization tests using Lim Benyesh–Melnick antisera pools [Reference Melnick23, Reference Melnick and Wimberly24] or EM and polyacrylamide gel electrophoresis [Reference Medici25, Reference Arcangeletti26].
RNA extraction and detection of NoV in clinical samples
For NoV detection, nucleic acids were automatically extracted by the NucliSens easyMAG instrument (bioMérieux, France) from 500 μl of stool suspension in a final volume of 25 μl. Following the manufacturer's instructions, detection of NoV RNA was performed using the RT Kit Plus (Nanogen Advanced Diagnostics, Italy) for reverse transcription (RT), and the NoV Oligomix Alert kit (Nanogen Advanced Diagnostics) for nested PCR (nPCR), targeting a 174–176 bp fragment of NoV RdRp, using CPE-RNA (Nanogen Advanced Diagnostics) as an internal RNA standard control.
Stool samples, faecal suspensions and first-round PCR products were stored at −20°C until use.
Nucleotide sequencing and molecular typing
Sequence analysis was undertaken on amplicons (327 bp in length), yielded with primer pair JV12/JV13 that target the ORF1 RdRp region A [Reference Medici22], from a set of NoV-positive specimens, selected according to the seasonal distribution of NoV infections.
For potential recombinant genomes of NoVs, and to exclude cases of mixed infection, the partial ORF1 RdRp and ORF2 capsid gene sequences along with the junction between RdRp and capsid (region A–C, 1110 bp) of a selection of strains representative of the RdRp lineages and sublineages were amplified by RT–PCR using primers JV12 and G2R1 [Reference Vinjé and Koopmans8, Reference Kojima14].
The Superscript One-Step RT–PCR for Long Templates kit (Invitrogen, USA) was used. Briefly, 5 μl extracted RNA was added to 12·5 μl of 2 × reaction mix, 4·5 μl H2O, 0·5 μl RT/Platinum Taq HiFi mix, containing 10 pmol of each primer. The reaction was performed in a PerkinElmer 9700 Thermal Cycler (Applied Biosystems, USA) with the following temperature conditions: 42°C for 60 min, 94°C for 2 min, 40 cycles at 94°C for 1 min, 50°C for 2 min and 68°C for 2 min, followed by 72°C for 10 min. Finally, amplicons of ORF2 region D (254 bp) were also produced from GII.4 strains using published primers and protocols [Reference Vinjé, Hamidjaja and Sobsey11].
The PCR products were electrophoresed on ethidium bromide-stained 2% agarose gel at 120 V for 40 min, visualized under UV light by GelDoc XR instrument (Bio-Rad Laboratories, USA), and purified using QIAquick Gel Extraction kit (Qiagen, Germany). Sequencing was performed using the PCR primers and BigDye Terminator Cycle Sequencing kit (Applied Biosystems) on an automated DNA sequencer (ABI 3730, PerkinElmer–Applied Biosystems, USA). All sequences were typed with the genotyping tool for NoVs (http://www.rivm.nl/mpf/norovirus/typingtool) [Reference Kroneman27].
Phylogenetic analysis of NoV GII.4
In order to investigate the genetic relationship among GII.4 strains, phylogenetic analysis was performed with the generated RdRp and capsid sequences for NoV GII.4 strains identified during the study period. The sequences were analysed using MEGA 5·0 software [Reference Tamura28]. Maximum composite likelihood was used as the substitution method, and the neighbour-joining method was used for construction of the phylogenetic tree. The reliability of the phylogenetic tree was assessed by bootstrap re-sampling over 1000 replicates. The BLAST web-based analysis tool with default values was used to find homologous hits in the sequence database (GenBank, NCBI). The nucleotide sequences have been deposited in GenBank under accession numbers KF743088–KF743123 (RdRp region), and KF743064–KF743087 (capsid region).
Statistical analysis
Statistical analysis was performed in order to compare the rates of detection of NoVs in different age groups and seasons, and to understand whether some NoV genotypes may preferentially infect some age groups, or whether patients with co-infections are more susceptible to some NoV strains. The χ 2 test was applied to the data, using GraphPad instat v. 3·05 for Windows (GraphPad Software, USA), with P < 0·05 considered to be significant.
RESULTS
Epidemiological features and NoV genotyping
NoV was detected as a single pathogen in 368 (12·98%, with a range from 9·8% in 2011 to 17·2% in 2008) cases whereas, 50 (1·8%), 13 (0·2%) and four (0·1%) samples had mixed infections with RV, AdV and enterovirus, respectively (Table 1). A total of 296 (68·04%) of the NoV-positive samples were from children aged <3 years (P = 0·0001) (Fig. 1). Of this group of patients, 46 (15·54%) children showed mixed infection with RV in 35 cases (52·23%), AdV in nine cases (13·43%) and enterovirus in two cases (2·98%). The monthly distribution plot showed that NoV was detected throughout the year, except in July 2007 and July–August 2011, with an increasing number of infections during the autumn–winter period (73·56%; 320/435) (P = 0·0001) (Fig. 2).

Fig. 1. Age distribution of children with norovirus-associated gastroenteritis (January 2007–December 2011).
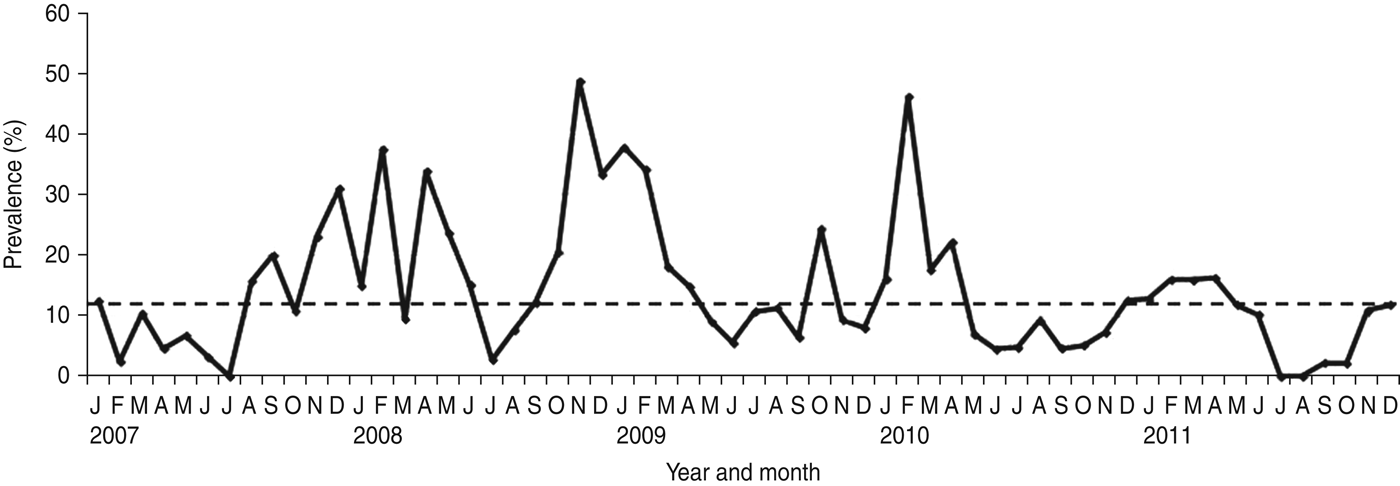
Fig. 2. Monthly distribution of sporadic cases of norovirus-associated gastroenteritis (January 2007–December 2011). The horizontal dotted line on the graph represents the median percentage of positive cases per month.
Table 1. Distribution of viruses detected in children with acute gastroenteritis between January 2007 and December 2011

NoV, Norovirus; RV, rotavirus; AdV, adenovirus; EV, enterovirus; ReoV, reovirus; HCaV, calicivirus observed by electron microscopy.
A total of 217 (49·88%) NoV strains were sequenced in the RdRp (region A). Seven typed NoVs (3·22%) belonged to GI, including four different genotypes (GI.P1, GI.P2, GI.P4, GI.P8), and 210 (96·77%) belonged to GII, of which 168 (80%) were characterized as GII.P4 (Table 2, Fig. 3). Within the GII.P4 genotype, four distinct variants (Yerseke 2006a, Den Haag 2006b, Apeldoorn 2008, New Orleans 2009), were identified. The remaining 42 (20%) GII NoVs included 19 (45%) GII.Pb, eight (19%) GII.P1, five (12%) GII.Pg, two (4·8%) GII.P2, two (4·8%) GII.P7, two (4·8%) GII.P16, two (4·8%) GII.Pe, one (2·4%) GII.P3, and one (2·4%) GII.P6.

Fig. 3. Norovirus polymerase genotypes revealed from 217 children with gastroenteritis (January 2007–December 2011).
Table 2. Number of norovirus genotypes detected in Parma, Italy, from 2007 to 2011
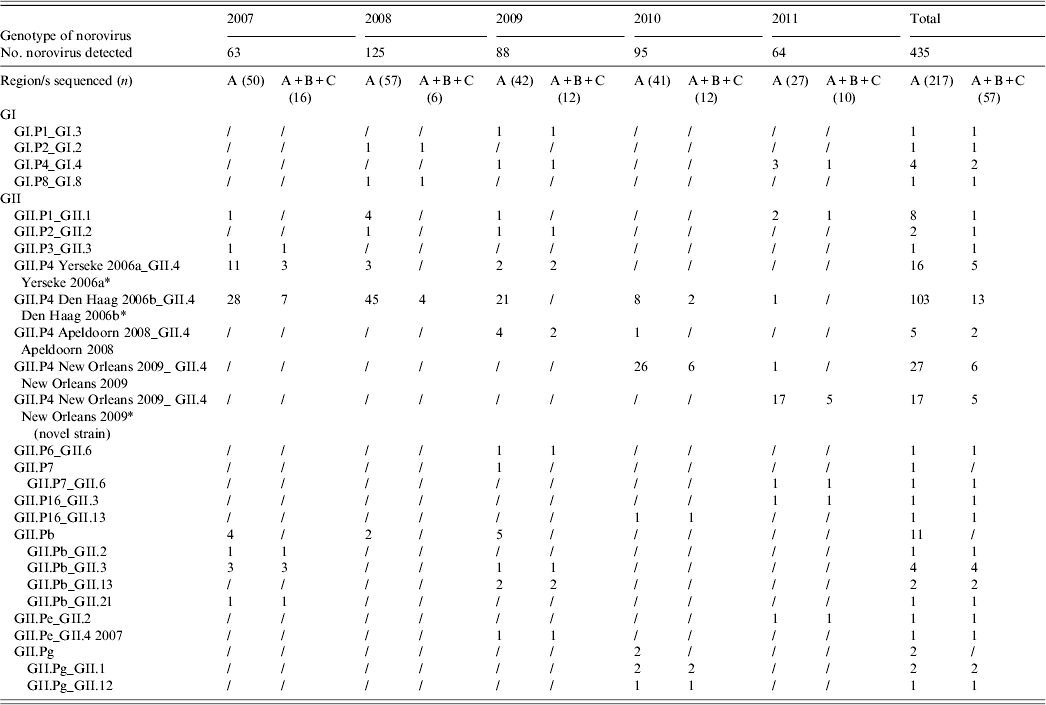
* Four GII.P4 Yerseke 2006a_ GII.4 Yerseke 2006a, 12 GII.P4 Den Haag 2006b_ GII.4 Den Haag 2006b, three GII.P4 New Orleans 2009_GII.4 New Orleans 2009 and two GII.P4 New Orleans 2009_ GII.4 New Orleans 2009 (novel strain) strains were also sequenced in the capsid region D.
Sequencing of the A–C genomic fragment of 57 strains allowed the identification of 17 potential NoV recombinants with a total of 12 different combinations of RdRp and capsid genotypes: GI.P1_GI.3, GII.P7_GII.6, GII.P16_GII.3, GII.P16_GII.13, GII.Pb_GII.2, GII.Pb_GII.3, GII.Pb_GII.13, GII.Pb_GII.21, GII.Pg_GII.1, GII.Pg_GII.12, GII.Pe_GII.2, and GII.Pe_GII.4 2007. For the other 40 strains, the capsid genotype matched the RdRp-based identification (Table 2). Multiple genotypes were found in co-infection with other enteric viral pathogens. Out of 19 genotypes identified in 67 mixed infections, eight were GII.P4 Den Haag 2006b, five were GII.P4 New Orleans 2009, two were GII.P4 Yerseke 2006a, and one each for GII.P4 Apeldoorn 2008, GII.Pb, GII.P1, and GII.P2. No statistical correlation was found between genotypes and co-infections. Similarly, different NoV genotypes were found in various age groups. The distribution in age groups of the NoV genotypes matched the distribution of NoV prevalences by age. A statistical correlation was observed between GII.P4 Den Haag 2006b NoV and age <3 years (P = 0·0078).
Phylogenetic analysis of GII.4
Phylogenetic clustering with GII.4 reference strain sequences from Noronet European genotyping tool displayed the genetic relationships among the 168 RdRp GII.4 genotypes detected in this study. In order to eliminate redundancy and prepare a readable figure, 36 sequences representative of the topology of branching of the RdRp GII.4 phylogenetic tree were used to prepare the tree in Figure 4 a.

Fig. 4. (a) Neighbour-joining phylogenetic trees based on partial nucleotide sequences of norovirus polymerase gene (284 bp, corresponding to nucleotides 4319–4602 referring to Lordsdale strain with GenBank accession number X86557) and (b) on partial nucleotide sequences of capsid C region (230 bp, corresponding to nucleotides 5102–5332 referring to Lordsdale strain with GenBank accession number X86557). The reference sequences were retrieved from the NoroNet database. Bootstrap values estimated with 1000 pseudo-replicate datasets, are indicated at each node.
The NoV GII.P4 strains grouped in two clades with different lineages and sublineages in the phylogenetic tree. One clade including the Italian NoVs belonged to GII.P4 Yerseke 2006a, Apeldoorn 2008 and New Orleans 2009 (bootstrap values 50–79%) and the second clade included the Italian GII.P4 Den Haag 2006b strains (bootstrap value 79%). The branching order showed that almost all the GII.P4 New Orleans 2009 strains detected in 2011 formed a close divergent lineage (bootstrap value 53%), that have a 4% nucleotide difference from the GII.P4 New Orleans 2009 strains. The 2011 GII.P4 strains were genetically highly homogenous (99% nucleotide identity to each other in the 1100 bp A–C genomic fragment).
The capsid phylogenetic tree of 24 NoV Italian GII.4 sequences identified in this study showed that the clustering patterns were consistent with those observed in the RdRp-based analysis with the divergent lineage of the GII.4 New Orleans 2009 strains detected in 2011 (bootstrap value 45%) (Fig. 4 b). This lineage was phylogenetically distinct also from the new GII.4 Sydney 2012 reference strain. In both the capsid regions C and D, the 2011 GII.4 viruses differed by 3–4% from the GII.4 New Orleans 2009 variant.
DISCUSSION
NoV is regarded as a major aetiological agent of sporadic diarrhoea in infants and young children [Reference Koopmans1–Reference Green, Knipe and Howley5]. During 5 years of surveillance for viral agents in sporadic cases of childhood acute gastroenteritis in Parma, Northern Italy, NoV was detected in 15·34% of children. The prevalence rates of NoV infection (with NoV as single pathogen) ranged between 9·8% in 2011 and 17·2% in 2008, revealing annual variations, as observed elsewhere [Reference Hoa16]. NoV infection occurred at higher frequency in children aged <3 years (68·04% in this study), an age-related pattern that has been reported in other studies, confirming the relevance of NoV as a cause of severe gastroenteritis requiring hospitalization in young children [Reference Bereciartu, Bok and Gómez29–Reference Junquera40]. In temperate climates NoV infections usually show a seasonal pattern with a peak during the cold months [Reference Mounts41–Reference Dey43]. In this study, seasonal peaks of NoV activity were revealed between January and February every year from 2007 to 2010, while in 2011 the activity was prolonged and a peak was reached in April. Moreover, a summer peak was observed in August in 2009. The peaks of NoV activity tended to overlap with or to anticipate the peaks of RV activity in all the surveyed years, except in 2011, when this pattern was inverted (data not shown).
Genotyping of the NoV strains identified during 2007–2011 revealed that different NoV genotypes did co-circulate during the study period and that several reported GII.4 variants were also detected in this cohort. The predominance of GII.4 NoVs (77·4%) is consistent with the literature [Reference Vinjé and Koopmans8, Reference Hoa16, Reference Siebenga17, Reference Bull20]. NoV GII.4 strains appear to evolve with genetic drift and shift mechanisms, with the periodic onset and rapid global spread of pandemic variants [Reference Vinjé and Koopmans8, Reference Hoa16, Reference Siebenga17, Reference Bull20]. This temporal pattern of evolution was also documented in our study. Variant GII.4 Yerseke 2006a was detected at a high frequency during 2007 and variant GII.4 Den Haag 2006b was predominant during 2007–2009, while variant GII.4 Apeldoorn 2008 was detected sporadically during 2009–2010. In 2010, variant GII.4 New Orleans 2009 emerged and quickly replaced, almost completely, the former variants. Strains belonging to the GII.4 New Orleans 2009 variant clustered along with strains of GII.4 Yerseke 2006a in the same lineage in the phylogenetic tree showing their close evolutionary relationships. The evolutionary trend analysis confirmed the continuous pattern of co-divergence of GII.4 variants, with GII.4 Den Haag 2006b linked closely to GII.4 Farmington Hills 2002, and the GII.4 variants US 95_96 and Yerseke 2006a related to the variants Apeldoorn 2008 and Hunter 2004. The variant New Orleans 2009 derived from the pandemic variant Yerseke 2006a and the new GII.4 variant Sydney 2012 share a common ancestor in the capsid gene with GII.4 variants Apeldoorn 2008 and New Orleans 2009. Interestingly in this study, a novel GII.4 strain, descending from GII.4 variant New Orleans 2009, was identified in 2011, showing that this variant is undergoing further diversification. This novel GII.4 strain differed by 4% nucleotides in the RdRp and by 3–4% nucleotides in both the C and D capsid regions from NoVs of GII.4 variant New Orleans 2009. The onset of this novel GII.4 strain was not associated with significant increased NoV activity, as the prevalence rates did not exceed the prevalence rates observed in the epidemic seasons 2007–2008, 2008–2009, and 2009–2010 (data not shown). This is not surprising as the onset and spread of novel GII.4 variants is not always related to increase in NoV activity [Reference Siebenga17]. Whether this novel GII.4 strain may represent a novel GII.4 variant should be assessed in larger and concerted epidemiological studies and with analysis of the data retrospectively. However, hospital-based surveillance studies can offer a reliable approach to monitor promptly the onset on novel NoV strains and variants in a settled population.
NoV RdRp genotypes GII.P16, GII.Pb, GII.Pe, and GII.Pg associated with multiple capsid types were identified in this study, suggesting the circulation of several recombinant strains. For these strains, SimPlot analysis on ORF1/ORF2 contigs confirmed the recombinant nature, with the breakpoint being located in the ORF1/ORF2 junction region (data not shown). NoV recombination has been described frequently, and the crossover points are usually located at highly conserved sequence stretches, such as the ORF1/ORF2 junction region [Reference Bull6]. In recent years, the recombinant NoV strains GII.Pb/Hilversum have been found to be common agents of gastroenteritis in Europe, chiefly in children. GII.Pb/Hilversum NoV strains share a common polymerase gene and display a variety of capsid genes, such as GII.1 (Hawaii), GII.2 (Snow Mountain), GII.3 (Toronto) and GII.4 (Lordsdale) [Reference Hoa16]. Interestingly, in the present study we observed 2010 NoV strains with either RdRp genotypes GII.Pe or GII.Pg apparently replacing NoV GII.Pb, which was common before 2010, suggesting a temporal shift. Many novel RdRp/capsid genotypes (GII.P7_GII.6, GII.P16_GII.3, GII.P16_GII.13, GII.Pe_GII.2, GII.Pe_GII.4 2007) were identified for the first time in sporadic cases of acute gastroenteritis in Italy, confirming that multi-target (ORF1- and ORF2-based) analysis is required in order to characterize properly NoV strains.
In conclusion, during a 5-year surveillance period, GII.4 were the predominant NoV strains in Northern Italy. Moreover, a candidate novel GII.4 variant was identified and a variety of NoV recombinants were detected for the first time in the country.
Since the epidemiology of NoV changes rapidly, enhanced monitoring of circulating NoVs will allow us to understand the current state of NoV infections in the population and support appropriate future public health intervention strategies against this important cause of sporadic childhood gastroenteritis.
ACKNOWLEDGEMENTS
This study was partly supported by the grants national project PRIN 2008 (20084F4P7X_004) granted by the Italian Ministry of Education, University and Research.
DECLARATION OF INTEREST
None.








