INTRODUCTION
The epidemiology of human cryptosporidiosis caused by Cryptosporidium (C.) hominis and C. parvum, and parasite species-specific risk factors have been well described [Reference Hunter1–Reference Chalmers4]. Two UK studies covering a 7-year period between them showed that nearly all typed cases were caused by C. hominis (54%) or C. parvum (45%). Other species and genotypes were found more rarely, being identified in the remaining 1% [Reference Chalmers3, Reference Chalmers4]. These species and genotypes which are unusual in the UK included C. meleagridis, C. ubiquitum, C. felis, C. canis, horse genotype, skunk genotype and novel genotypes. C. hominis and C. parvum were especially prevalent in children, and there was no difference in the distribution of these two species in immunocompetent and immunocompromised patients [Reference Chalmers3, Reference Chalmers4]. Significant risk factors, identified in a case-control study of sporadic cases, for C. hominis infection included area of UK residence, travel outside the UK, and changing nappies [Reference Hunter1]. For C. parvum, significant risk factors were contact with farm animals [Reference Hunter1], and in a study where 84% of cases were C. parvum, increasing volume of tap water drunk and short visits to farms [Reference Goh5]. Little is known about the epidemiology of infections with other Cryptosporidium spp. in the UK, although individual case reports and descriptions of affected patients have been published [Reference Robinson, Elwin and Chalmers6–Reference Pedraza-Diaz8]. In contrast, epidemiological studies of cryptosporidiosis in developing countries have involved more cases with other species and genotypes. In a study of children in a shanty town in Lima, Peru no difference in associations between drinking-water supply, sanitation, presence of animals or socioeconomic status and unusual cryptosporidiosis compared to C. hominis were found [Reference Xiao9]. In the same setting, Cama and colleagues [Reference Cama10, Reference Cama11] described Cryptosporidium infections in children and HIV-positive adults and found a high frequency of zoonotic species (C. meleagridis, C. felis, C. canis) in an urban area leading to their suggestion that there may be anthroponotic transmission of species with seemingly animal origins. It is clear from the available literature that, like C. hominis and C. parvum, C. meleagridis, C. canis and C. felis are found worldwide, but appear to be more common in developing countries [Reference Xiao9–Reference Gatei12]. In contrast, with the exception of three children in Nigeria [Reference Molloy13], infections with C. ubiquitum (syn. cervine genotype [Reference Fayer, Santin and Macarisin14]) have only so far been reported in developed nations [Reference Ong15–Reference Davies17]. It is therefore important for us to understand the occurrence and epidemiology of these species and genotypes in the UK as they have a higher prevalence elsewhere and are deemed to be pathogenic to humans. This information will help interpret risk presented by, for example, their finding during water monitoring, and in understanding potential reservoirs of these infections.
To investigate the epidemiology of sporadic, unusual cases in England and Wales we compared their demographics and exposure data with those of C. hominis and C. parvum cases from the national dataset, from the same 9-year period, followed by comparison of data between the unusual species cases.
METHODS
Species and genotype characterization
Unusual Cryptosporidium species and genotypes were identified routinely by the UK Cryptosporidium Reference Unit (CRU) under the national typing scheme between January 2000 and December 2008 [Reference Chalmers3, Reference Chalmers4] as isolates having PCR–RFLP profiles of the Cryptosporidium oocyst wall protein (COWP) gene using RsaI [Reference Spano18], other than those matching C. parvum or C. hominis. Additionally, isolates from cases confirmed by microscopy but not amplifiable using the COWP locus were investigated further. First, DNA was re-amplified and analysed using a small sub-unit (SSU) rDNA PCR–RFLP using SspI and VspI [Reference Xiao19, Reference Jiang, Alderisio and Xiao20] and then identity was confirmed by bi-directional PCR sequence analysis (Geneservice, UK: ABI 3730xl; in-house: CEQ8000, Beckman Coulter, UK). The exception to this routine test algorithm was that the identities of C. canis and a novel genotype were established using an HSP70 PCR [Reference Morgan21] as amplicons suitable for further analysis could not be generated from the first choice loci. Representative sequences were deposited into GenBank using the online submission protocol.
Enhanced surveillance of unusual cases
An unusual case was defined as a person who had submitted a stool sample for diagnosis of gastrointestinal symptoms and had Cryptosporidium oocysts identified routinely as a species other than C. parvum or C. hominis. To obtain descriptive epidemiological data about each case, a questionnaire was sent to the lead consultant of the local Health Protection Team. Data from initial trawling questionnaires, based on routine investigation of sporadic cases [Reference Bouchier22] containing personal details and basic exposure data on known Cryptosporidium risk factors, were transferred to an enhanced questionnaire. This was augmented with clinical data from the patient on immune status, diagnosis of other gastrointestinal pathogens, and further animal exposures. Case occupation, derived from an open question, was also recorded and grouped according to risks of transmission, i.e. those that worked on a farm, with food, with children (e.g. carers at home, nursery staff, school staff), or had close contact with other people (e.g. care workers, hospital) were categorized as ‘high risk’. Other occupations were categorized as ‘low risk’.
Data analysis
Exposure data were entered and stored in a Microsoft Excel spreadsheet with the Cryptosporidium typing results and transferred to Epi Info (version 6, Centers for Disease Control and Prevention, USA) for analysis. Demographic and epidemiological data for all unusual cases were compared to those for C. parvum and C. hominis cases and to each other using χ2 test to compare patient's sex, seasonal quarter of specimen receipt, foreign travel and immune status (the latter for cases occurring during 2000–2003 only). Mann–Whitney U test was used to compare case age. To investigate foreign travel case ages were arranged into groups stratified by 10-year bands. Basic statistical data, describing significance of differences between groups for each variable are given in Tables 1 and 2 and more detailed analyses are described in the text. Analysis of data within the unusual case group was only carried out with C. meleagridis, C. felis and C. ubiquitum as separate groups due to numbers of other species/genotypes being very small. Demographic and clinical data on the very small numbers of other species or genotypes are provided in Table 2.
Table 1. Comparison of unusual cases with Cryptosporidium hominis and C. parvum cases
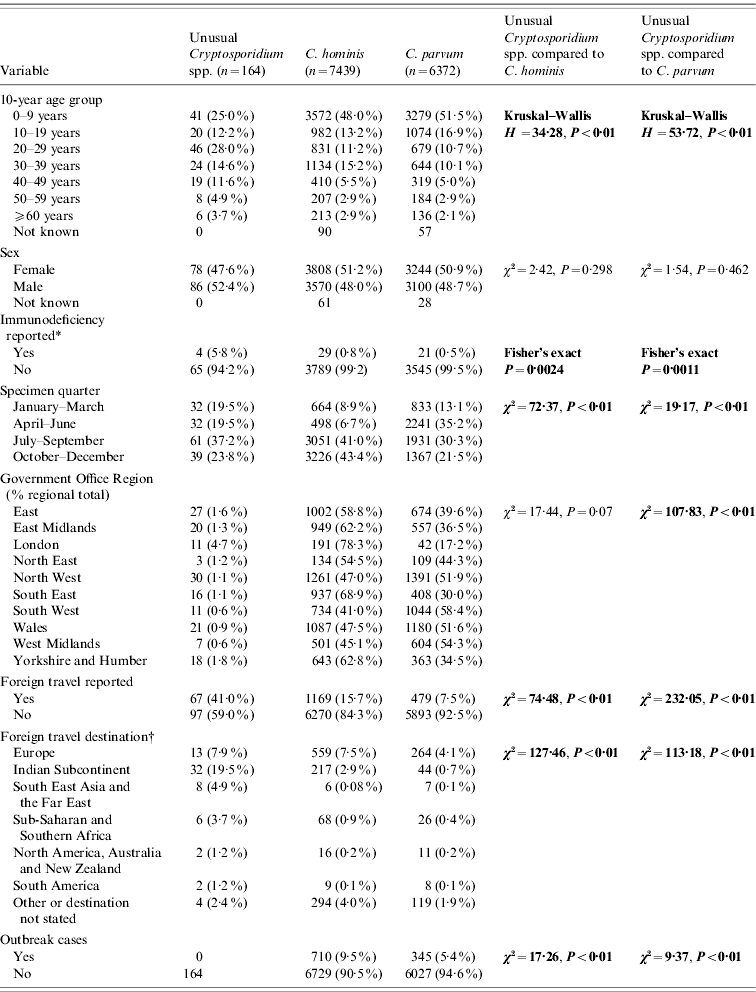
* Data from 2000 to 2003 only.
† Yellow book list of countries by continental group (National Travel Health Network and Centre, NaTHNaC, HPA UK).
Significant statistics shown in bold.
Table 2. Description of all unusual cases and comparison of variables between Cryptosporidium meleagridis, C. felis and C. ubiquitum
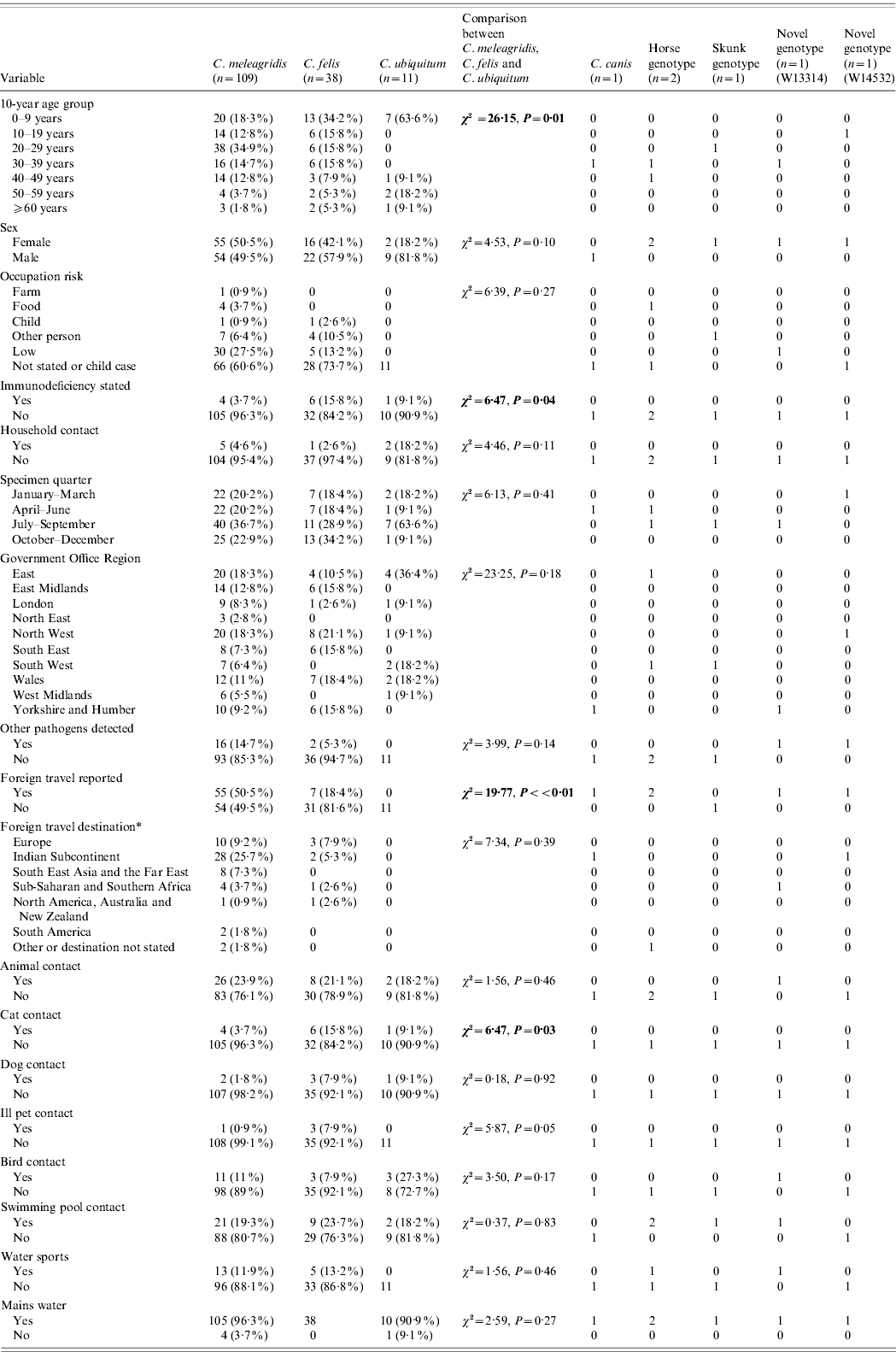
* Yellow book list of countries by continental group (National Travel Health Network and Centre, NaTHNaC, HPA UK).
Significant statistics shown in bold.
RESULTS
Species and genotype identification
Of 14 469 confirmed Cryptosporidium isolates from individual patients (one per patient) submitted to CRU during the 9-year period, 164 (1·1%) were unusual Cryptosporidium species or genotypes, 7439 (51·4%) were C. hominis, 6372 (44·0%) C. parvum, 51 (0·4%) both C. hominis and C. parvum, and 443 (3·1%) were not typable. Of the unusual cases, 109 were C. meleagridis, 38 C. felis, 11 C. ubiquitum, two horse genotype, two novel genotypes and one each of C. canis and skunk genotype. One C. hominis monkey genotype case and 37 C. cuniculus cases were identified by a look-back exercise during 2007 and 2008 using a different test algorithm, targeting species/genotypes closely related to C. hominis and the data presented elsewhere [Reference Chalmers23]; these cases have been removed from this dataset as they were not identified routinely.
Representative sequences have been deposited into GenBank under accession numbers EU437414-EU437418, HM191258-HM191264 and HM485431-HM485435.
Enhanced surveillance of unusual cases of cryptosporidiosis and comparison with C. hominis and C. parvum
Completed questionnaires were returned from 103/164 (63%) unusual cases, although a basic dataset of demographic data was available for all cases from the specimen request forms (Tables 1 and 2). There were no differences in age group or sex distribution between cases who did and did not return a questionnaire (χ2=3·28, P=0·77; χ2=0·42, P=0·52, respectively). Regional variation approached significance (χ2=16·73, P=0·053) which may be due to local differences in case follow-up procedures and therefore independent of the case. Specimen date was incomplete for 12/164 (7·3%) unusual cases and for 882/13 811 (6·4%) C. parvum and C. hominis cases.
Almost half the unusual cases were female (47·6%) which is comparable with cases of C. hominis and C. parvum. Unusual case age ranged from 0 to 82 years (median 24 years) and was significantly older than cases with C. hominis (median 10 years, χ2=34·28, P<0·01) or C. parvum (median 9 years, χ2=53·72, P<0·01). Only 17% of cases with unusual species were aged ⩽5 years compared to 35% of C. hominis cases and 37% of C. parvum cases.
Data about patient immune status were collected for C. parvum and C. hominis up to the end of 2003: being immunocompromised was significantly more frequent in unusual cases (4/69, 5·8%) compared to C. hominis cases (29/3818, 0·8%) (Fisher's exact P=0·002) and C. parvum cases (21/3566, 0·6%) (Fisher's exact P=0·001) during this period.
Unusual species were recorded throughout the year, but the highest proportion of specimens was received between July and September (61/164, 37·2%). This is significantly different from C. hominis samples, most of which were received between October and December (3226/7439, 43·4%; χ2=72·37, P<0·01) and C. parvum samples, most of which were received between April and June (2241/6372, 35·2%; χ2=19·17, P<0·01).
Samples were submitted from cases living in all ten Government Office Regions of England and Wales. There were regional differences in the distribution of unusual cases compared to C. hominis and C. parvum. The highest proportion of unusual cases was in Yorkshire and the Humber (18/1024, 1·8%); the highest proportion of C. hominis was in the South East (937/1361, 68·8%) and C. parvum in the South West (1044/1789, 58·4%). Data from the London region were omitted from regional analysis as few samples were sent for typing.
Foreign travel was reported by 67/164 (41·0%) of the unusual cases which is significantly higher than reported for C. hominis (1169/7439, 15·7%; χ2=74·48, P<0·01) and C. parvum (479/6372, 7·5%; χ2=232·05, P<0·01) infections. Of the 67 unusual cases that indicated travel, 65 (97·0%) stated the destination. These were predominantly the Indian subcontinent (India, n=24; Pakistan, n=3; Nepal, n=5) (total 32/65, 49·2%). Destination was stated by 1082/1169 (92·6%) C. hominis cases and by 428/479 (89·4%) C. parvum cases. Unusual cases were more likely to have travelled to the Indian subcontinent than returning travellers with C. hominis (223/1082, 20·6%; χ2=29·08, P<0·01) or C. parvum (51/428, 11·9%; χ2=56·00, P<0·01). Returning travellers with C. hominis or C. parvum were more likely to have come from Europe than unusual cases (823/1510, 54·5%; χ2=29·77, P<0·01) (Table 1). Of the unusual cases, those who had travelled abroad were more likely to have used a swimming pool (22/67, 32·8%), than those who had not travelled abroad (14/97, 14·4%; χ2=7·79, P<0·01).
None of the unusual cases which we identified through routine testing were part of a recognized outbreak of cryptosporidiosis compared to 710/7439 (9·5%) C. hominis cases (χ2=17·26, P<0·01) or 345/6372 (5·4%) C. parvum cases (χ2=9·37, P<0·01).
Comparisons between C. meleagridis, C. felis and C. ubiquitum
When case data for individual unusual species were compared (Table 2), no difference in the sex distribution was observed (χ2=4·53, P=0·1) but there was a notable excess of males in C. ubiquitum cases (9/11, 81·8%) compared to all other unusual cases (χ2=3·71, P=0·05). Case ages varied according to species (χ2=26·15, P=0·01). C. ubiquitum cases peaked in young children (mode 1 year), C. meleagridis and C. felis cases were spread across all ages but C. meleagridis peaked in young adults (mode 24 years), with significantly more aged 20–29 years than any other unusual species (χ2=8·55, P=0·003). C. felis peaked in children with 34·2% in those aged <9 years (mode 1 year).
In adult cases, occupation group was not significantly associated with any unusual species (χ2=6·39, P=0·27); most occupations were categorized as low risk. Where the case was a child, parental occupation group was not linked to a specific species or genotype (χ2=11·77, P=0·30).
Being immunocompromised was more common in cases with C. felis (6/38, 15·8%, Fisher's exact P=0·02) than other unusual cases. Where stated, HIV infection was the most common cause of immunodeficiency in the unusual cases (6/9 cases, 66·7%).
The presence of diarrhoeal illness in household contacts was not associated with any particular unusual Cryptosporidium spp. (χ2=1·56, P=0·46).
No significant difference in seasonal quarter of specimen receipt between unusual species was evident (χ2=6·13, P=0·41).
There were no significant differences in the regional distribution of unusual species (χ2=23·25, P=0·18), although the East of England had more C. ubiquitum cases (4/11, 36·4%) than any other region and the North West had more C. felis cases (8/38, 21·1%).
The presence of other gastrointestinal pathogens was not associated with any single unusual species of Cryptosporidium (χ2=3·99, P=0·14). Co-infections were reported with Campylobacter spp. (n=14), Escherichia coli O157 (n=1), Salmonella sp. (n=1) and one case with multiple co-infections (Campylobacter spp., Entamoeba coli, Giardia spp.). Of the 15 unusual cases with Campylobacter spp., 13 were C. meleagridis cases (86·7%).
Travel abroad varied in the unusual cases (χ2=19·77, P<0·01) and in particular was undertaken more frequently by cases infected with C. meleagridis (55/109, 50·5%) than with other unusual Cryptosporidium spp. infections (7/49, 14·3%) (χ2=18·43, P<0·01). None of the 11 cases with C. ubiquitum had travelled. Where destination was stated more cases with C. meleagridis had travelled to the Indian subcontinent (28/54, 51·9%) than had C. felis cases (2/7, 28·6%) but this was not a significant difference (Fisher's exact P=0·23). Foreign travel was reported by all age groups (χ2=12·51, P=0·05) and peaked in the 20–29 years group. The choice of destination (where stated) was significantly linked to case age group (χ2=47·59, P<0·01); 15/30 (50%) travellers to the Indian subcontinent were aged 20–29 years (χ2=5·26, P=0·02).
Contact with any animals was not associated with infection with an unusual Cryptosporidium sp. (χ2=0·27, P=0·87). However, contact with cats was reported by significantly more C. felis cases (6/38, 15·8%) than cases with other unusual species (5/120, 4·2%, Fisher's exact P=0·02). Contact with dogs was not associated with an unusual species (χ2=0·18, P=0·92). Contact with an unwell pet was associated with an unusual species (χ2=5·87, P=0·05), with three of the four unusual cases who reported contact with an unwell pet identified as having C. felis, all of whom specifically reported kittens or cats that were unwell (Fisher's exact P=0·04). The fourth unusual case had C. meleagridis and reported contact with an unwell dog, a host from which C. meleagridis has been previously reported [Reference Hajdusek, Ditrich and Slapeta24]. Contact with birds was not associated with infection with an unusual species (χ2=3·50, P=0·17).
The use of swimming pools (χ2=0·37, P=0·83) or participation in water sports (χ2=1·56, P=0·46) were not associated with infection with an unusual species. Twenty percent of unusual cases reported contact with swimming pools which is less than that reported by 790 sporadic C. hominis and C. parvum cases (44·8%) during a 3-year study in the UK [Reference Chalmers4].
Drinking-water source (whether or not mains water was consumed) was not associated with any particular unusual species (χ2=2·59, P=0·27); however, 5/158 (3·2%) cases consumed water from a private supply.
DISCUSSION
Enhanced surveillance of unusual cases of cryptosporidiosis has provided insight into the epidemiological and behavioural factors associated with risk of infection with unusual Cryptosporidium species or genotypes in the population of England and Wales. The median age of unusual cases was 24·5 years, higher than that of C. hominis and C. parvum cases, and the age distribution showed a much lower proportion of cases aged <5 years than C. hominis and C. parvum cases. It is probable that behavioural factors are important here; for example foreign travel, especially to developing countries in the Indian sub-continent, is associated with unusual infections, particularly C. meleagridis, and such travel is more often undertaken by those aged 20–29 years than any other age group [25]. Although it could be argued that foreign travel may be used as a selection criterion for primary testing for Cryptosporidium spp., thus increasing detection rates in this age group, the unusual case-case comparison showed that the age distribution was unique to C. meleagridis in this dataset, and is unlikely to be a surveillance artefact. Unusual infections have been reported to occur in residents of developing countries such as India and elsewhere in Asia, where they may be endemic [Reference Gatei12, Reference Gatei26]. It is not known whether prior exposure to C. parvum or C. hominis is cross-protective against further illness caused by a heterologous isolate [Reference Chappell27], so the risk of infection for travellers to these destinations may not be mitigated by previous domestically acquired infection.
Being immunocompromised was associated with infection with an unusual species or genotype, but it is possible that our data are biased by better information on immune status actively sought for the unusual cases than that received passively for C. hominis and C. parvum cases on the routine genotyping request form.
When date of receipt of unusual case samples was compared with C. hominis and C. parvum differences were observed. These contrast to the well documented albeit recently reduced ‘spring peak’ [Reference Lake28] of C. parvum cases and the post-summer holiday period peak in the last quarter of the year of C. hominis cases [Reference Chalmers3]. Unusual cases peaked in the July–September period which may reflect slightly earlier travel dates, or seasonal exposure to other unknown risks. Specimen date was incomplete for a substantial number of cases; therefore seasonality was measured using date of receipt in our laboratory. Our previous experience using both specimen and receipt dates has shown that the latter differs from the former by an average of 5 days [Reference Chalmers3] which would not significantly affect seasonality reported here.
When the demographics and risk factors for specific unusual species or genotypes were compared an excess of young male cases of C. ubiquitum was seen which cannot be explained by data collected and may warrant further investigation. Most cases of C. meleagridis occurred in young adults, although in this scenario, age is probably acting as a confounder for foreign travel as discussed above.
In contrast none of the 11 cases with C. ubiquitum had travelled abroad and this suggests that C. ubiquitum in humans may be indigenous to the UK, which is supported by frequent identification in sheep and during environmental investigations here [Reference Elwin and Chalmers29–Reference Chalmers31]. Elsewhere C. ubiquitum has been found in a wide variety of hosts [Reference Fayer, Santin and Macarisin14], but in the UK sheep are a likely source of environmental contamination due to their high density [Reference Robertson32]. However, the number of human cases with C. ubiquitum is small and its pathogenicity is not known.
The association between cats and infection with C. felis is perhaps unsurprising as this is the predominant species in cats [Reference Santin, Trout, Fayer and Xiao33]. Although contact with companion animals has not been associated with cryptosporidiosis in epidemiological or microbiological studies [Reference Hunter1, Reference Smith34], individual infection risks may be present and hygiene advice should be followed. Since Cryptosporidium infection is by the faecal–oral route, either directly or indirectly, the most appropriate intervention is thorough hand-washing with soap and warm running water before preparing and eating food, after handling raw food, after going to the toilet or changing a baby's nappy, after working, feeding, grooming or playing with pets and other animals [Reference Smith34].
Within unusual cases, concomitant infection with Campylobacter was commonly reported. This concurs with previous findings in developed and developing nations [Reference Duke35, Reference Cama36] (CRU, unpublished observations). Campylobacter is the most commonly detected bacterial gastrointestinal pathogen in the UK [37] and is routinely sought in addition to Salmonella spp., Escherichia coli O157 and Shigella spp. in the diagnosis of gastrointestinal infections in the UK [38]. Conversely, the diagnosis and reporting of parasites such as Giardia spp. and Entamoeba spp. are often limited by selective testing of patients who have travelled abroad [38]. Nevertheless, the potential association between C. meleagridis and Campylobacter spp. is intriguing as both species have either recently or in the past been associated with poultry [Reference Ryan, Xiao, Fayer and Xiao39, Reference Arsenault40]. However, no significant association between bird contact and infection with C. meleagridis was established from the data presented here.
No unusual species or genotypes were the cause of a recognized outbreak of cryptosporidiosis through the routine test algorithm described here; however, it should be noted that C. cuniculus (known at the time as the rabbit genotype) was identified by CRU as the causative agent of a waterborne outbreak in 2008 through a separate test algorithm using SSU rDNA PCR–RFLP as the first-line test [Reference Chalmers41].
This study describes for the first time the epidemiology and risk factors for infection with unusual Cryptosporidium spp. in a developed country. Some of the unusual species are probably indigenous to the UK while others more likely to be acquired abroad. To properly understand the infectivity and pathogenicity of different Cryptosporidium spp. and genotypes more information is needed about the spectrum of illness of all causes of cryptosporidiosis and should therefore be the focus for future work of the kind presented here.
ACKNOWLEDGEMENTS
We are grateful to the Health Protection Units in England and Wales, who provided the epidemiological data presented here. We also thank Dr Brendan Mason for his helpful comments on our manuscript and the laboratory support and assistance of Anne Thomas, Nigel Crouch, David Gomez and Rachael Seymour.
DECLARATION OF INTEREST
None.




