INTRODUCTION
During the 2001 anthrax attacks in the USA, inhalational anthrax developed in 11 persons; five of whom died from their illness [Reference Jernigan1]. For eight of the 11 patients, Gram-positive rods (GPR) were initially identified from routine diagnostic blood cultures in <24 h from the time the patient's blood was drawn and the cultures inoculated. Genus and species-specific identification took several more days of additional testing of the bacterial isolates [Reference Jernigan2, Reference Griffith3]. The eleventh individual who developed inhalational anthrax was a 94-year-old Connecticut resident who was intubated for mechanical ventilation in the 2-day interval between initial identification of GPR growth in blood cultures and additional laboratory results specifically suggesting B. anthracis. According to 2001 Connecticut reporting requirements, the hospital laboratory notified the Connecticut Department of Public Health (CDPH) only after B. anthracis was suspected; CDPH staff were unable to interview the patient who soon died [Reference Griffith3].
Most cases of inhalational anthrax will develop clinical symptoms within 2–14 days after exposure, more than 25% within 7 days [Reference Meselson4]. Thus, it is critical to identify exposures as soon as possible in order to minimize the public health impact by reduction of exposure to contaminated areas, chemoprophylaxis of exposed individuals, early recognition and treatment of additional persons with disease to reduce the risk of death, and development of risk communication messages for the public. In a large-scale inhalational anthrax event, each day detection is delayed can result in thousands of additional deaths and millions of dollars in additional expenditures [Reference Kaufmann, Meltzer and Schmid5]. On a smaller scale during the anthrax attacks in 2001, timely chemoprophylaxis probably prevented nine additional cases among exposed persons [Reference Brookmeyer and Blades6]. One approach to early detection of a mass anthrax exposure event is to identify the initial case(s) of confirmed anthrax as rapidly as possible. Based on our experience in 2001, we reasoned that by establishing real-time, telephone-based surveillance for the occurrence of the earliest specific diagnostic finding [GPR identified by Gram stain of blood or cerebrospinal fluid (CSF) culture], we could shorten the time period for alerting public health authorities and for confirmation that an isolate was anthrax by 2–3 days.
Beginning in March 2003, Connecticut laboratories were required to report by telephone all GPR isolates detected within 72 h of culture inoculation from blood or CSF to the CDPH [Reference Begier7]. In 2004, the time-frame for reporting was reduced to positive growth within 32 h to reduce reporting of culture contaminants without significantly reducing sensitivity to detect anthrax or clostridial infections [8]. Although blood cultures from all recent anthrax cases in the USA grew within 24 h of collection, we chose 32 h to ensure that laboratories without routine night-shift blood-culture readings would report cultures that may have turned positive during the night. Surveillance objectives include: (1) sensitive and timely detection of anthrax septicaemia or meningitis independent of clinician or laboratory recognition; (2) round-the-clock laboratory reporting of an indicator of possible biological terrorism; and (3) surveillance for invasive Clostridium spp. infections of public health importance [Reference Jones9–Reference Fischer11].
While local, state, and federal agencies in the USA continue to research and implement costly systems to detect intentional disease outbreaks sooner [Reference Bradley12–16], we report on 4 years of experience with Connecticut's GPR surveillance system. This system underwent a comprehensive evaluation using the Centers for Disease Control and Prevention (CDC) guidelines for evaluating public health surveillance systems [17] after a year of operation in 2004; selected findings were published [Reference Begier7]. Here, we report on information obtained from the system since that time, examine the system in the context of recent anthrax reports, and follow-up on key aspects of the prior evaluation. Specifically, we describe the epidemiology of GPR bacteraemia and septicaemia from reporting those GPR isolates from blood or CSF identified within 32 h of inoculation [Reference Begier7, 8], examine the frequency of randomly occurring clusters of GPR illness, and report on the hypothetical impact of this system on the timeliness of initiation of investigation of the one reported case of inhalational anthrax in the USA since 2001 [18].
METHODS
Each of Connecticut's 34 clinical laboratories processed blood cultures according to their routine diagnostic protocols. Ideally, blood specimens drawn for microbial culture were obtained from at least two blood draws from separate venepuncture sites. These blood draws may have been performed simultaneously or over a short period of time (e.g. within a few hours). Blood from each blood draw typically was inoculated into two bottles (one primed to promote aerobic and one anaerobic bacterial growth). A blood culture may have consisted of a single bottle for a paediatric blood draw due to volume constraints [19]. Culture bottles were incubated and monitored for growth in the clinical laboratory. Immediately upon recognition of growth, laboratories routinely conducted a Gram stain to determine what type of organism was growing before initiating additional tests to determine the genus and species. If a GPR was identified, a telephone call was made to the CDPH. CDPH staff then obtained information on the number of blood-culture bottles and which were growing GPR and made a determination as to whether immediate clinical follow-up was needed.
In general, immediate clinical follow-up was conducted for all CSF reports and for blood cultures when multiple bottles from a single patient yielded an isolate within 32 h of inoculation [Reference Begier7, 8]. More specifically, the algorithm used to determine whether a report required immediate clinical follow-up to rule out anthrax or to identify unusual cases of clostridial sepsis was as follows (Fig. 1). If one aerobic bottle was positive, aerobic follow-up was conducted when the patient's chief complaint as reported by the laboratory was suggestive of inhalational anthrax (i.e. pneumonia, sepsis) or if no chief complaint was available. If multiple aerobic bottles were positive, clinical follow-up was conducted regardless of chief complaint.
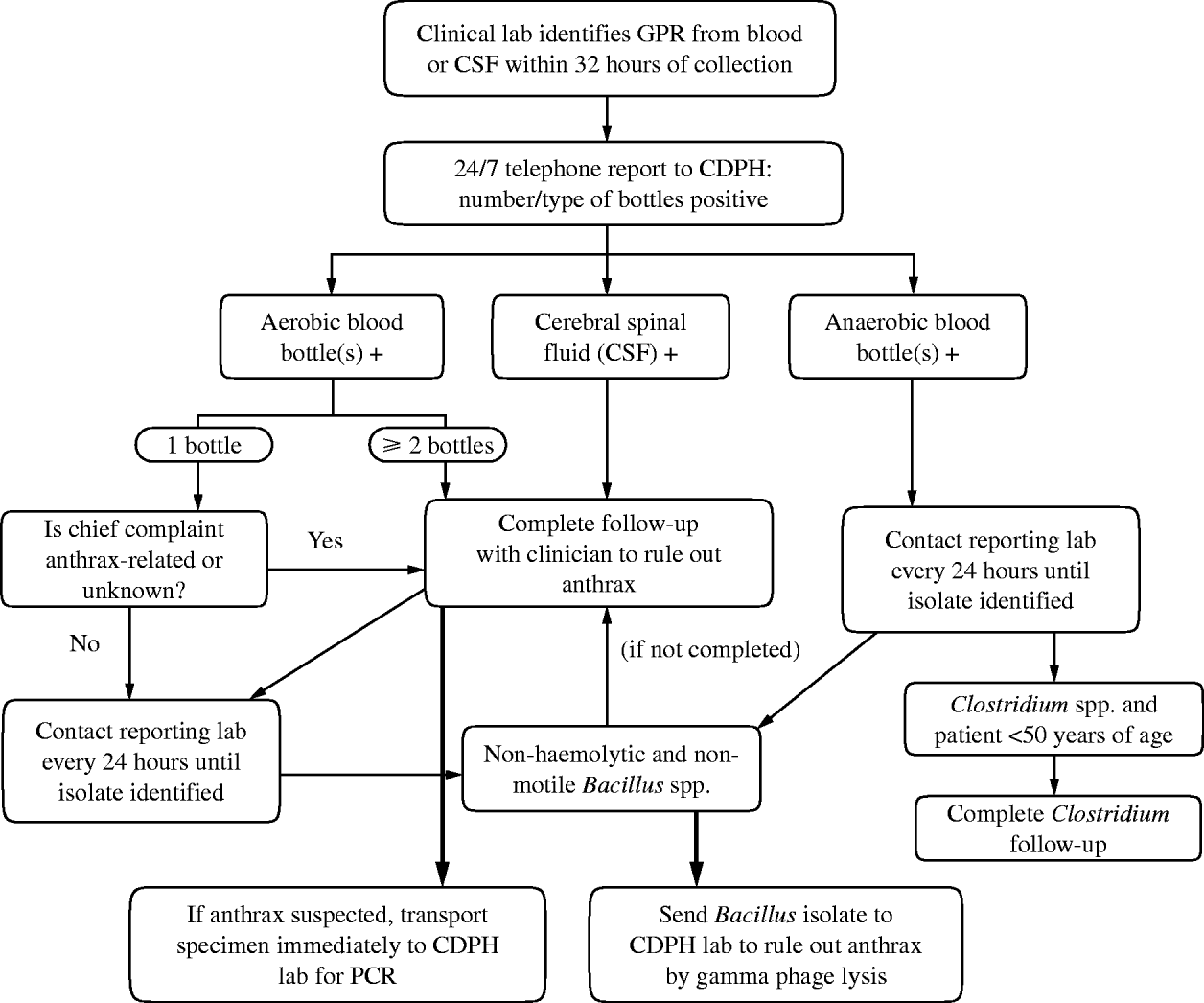
Fig. 1. Gram-positive rod (GPR) surveillance algorithm, Connecticut, 2003–2006. CDPH, Connecticut Department of Public Health; PCR, polymerase chain reaction.
Clinical aerobic follow-up involved physician contact to assess whether the patient's illness resembled inhalational anthrax (e.g. presence of respiratory symptoms, widened mediastinum on chest radiograph, or sepsis), whether the physician was considering anthrax as a diagnosis, the patient's clinical status, and any recent travel or possible occupational exposure history. If clinical follow-up determined that there was the possibility of anthrax infection, the isolate would be immediately transported to the CDPH laboratory for anthrax-specific testing. If only anaerobic bottles were positive, clinical follow-up was delayed until GPR genus was available from the reporting laboratory. In subsequent laboratory work-up of the isolate, if the isolate was a non-haemolytic, non-motile Bacillus spp., clinical aerobic follow-up, as described above, was conducted. If the isolate was identified as a Clostridium spp. and the patient was aged <50 years, clinical information (e.g. history of tissue allografts, injection drug use, recent medical abortion, or underlying conditions that may pre-dispose a patient to Clostridium spp. infections) was obtained to rule out an unusual clostridial event.
Daily laboratory follow-up was conducted for all isolates until at least genus identification. Mailed reports (including faxed reports) that were not preceded by a telephone report were followed up by CDPH staff by contacting the reporting laboratory to determine the genus and species of bacteria isolated. Laboratory staff were reminded of telephone reporting requirements. Any Bacillus spp. isolates for which clinical follow-up did not raise suspicion of anthrax, but were found by the clinical laboratory to be non-motile and non-haemolytic, were subsequently forwarded to the CDPH laboratory to rule out B. anthracis by gamma-phage lysis. Annual laboratory audits were conducted for Bacillus spp. and Clostridium spp. isolates to determine reporting completeness.
To increase the ability of the surveillance system to detect large-scale outbreaks of illness, CDPH staff regularly reviewed GPR reports to identify days on which there was a significant cluster of isolates and reviewed these cases to determine if an outbreak of specific GPR infection was occurring. A significant cluster of illness was defined as four or more GPR reports on any calendar day. Identification of a cluster resulted in enhanced clinical and laboratory follow-up (regardless of where the report would fall in the follow-up algorithm) to determine if any similarity existed among case reports. Follow-up of individual reports continued until adequate information was collected to rule in/out a point-source outbreak. An evaluation of the adequacy of the cluster definition was performed.
Connecticut has 34 clinical laboratories statewide and a population of 3·4 million people; the workload to maintain this system required less than the equivalent of one full-time employee, with 20% of work done during off hours [Reference Begier7]. Connecticut has a rotational after-hours on-call system staffed by senior epidemiologists that handled the after-hours reports.
Using Connecticut's algorithm for reporting, follow-up and confirmation of GPR infection, a simulation was conducted to assess whether Connecticut's GPR surveillance system would have detected anthrax more quickly than in the 2006 Pennsylvania inhalational anthrax case [18].
RESULTS
From 1 March 2003 to 31 December 2006, 1141 GPR isolates meeting Connecticut's reporting requirements were identified through reporting and subsequent laboratory audits. Only seven CSF isolates occurred (five Bacillus spp., one Listeria, one Corynebacterium) and are excluded from analysis. Overall annual average incidence of GPR blood isolates was 8·6/100 000 population (range 7·3–9·9); Bacillus spp. and Clostridium spp. isolates had overall average annual incidence rates of 4·9 and 1·9/100 000 population, respectively (Table 1).
Table 1. Gram-positive rod (GPR) bacterial isolates from blood culture and annual incidence, Connecticut, 2003–2006
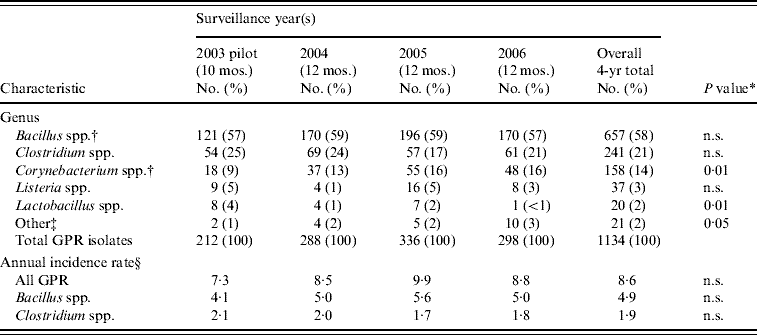
* χ2 test for linear trend.
† No Bacillus anthracis or Corynebacterium diphtheriae were reported.
‡ Other category includes Eubacterium spp. (6), Propionibacterium spp. (3), Brevibacterium spp. (2), Actinobaculum spp. (1), Actinomyces spp. (1), Bifidobacterium spp. (1), Erysipelothrix spp. (1), and isolates with unknown/uspecified genus (6).
§ Annual incidence rate for 2003 estimated using mean of January 2004–2006 and February 2004–2006 as 2 months were missing in 2003; number of GPR reports did not differ significantly for either month across the three surveillance years.
Table 2 gives selected attributes of the surveillance system and how they have changed over time. Completeness of reporting improved significantly. Based on audits of 97% of Connecticut's 34 clinical laboratories, unreported isolates of Bacillus spp. and Clostridium spp. decreased over time, accounting for 27% of these genera in 2003 but only 7% in 2006 (P<0·01). Telephone reporting increased from 75% in 2003 to >92% in 2005 and 2006 (P<0·01). About one third of these telephone reports were made outside of normal business hours.
Table 2. Reporting system attributes and trends in reporting of Bacillus spp. and Clostridium spp. isolates from blood culture, Connecticut, 2003–2006
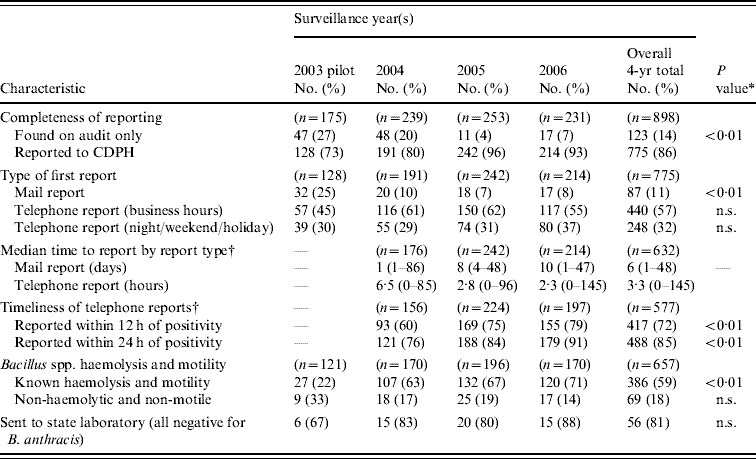
CDPH, Connecticut Department of Public Health.
* χ2 test for linear trend.
† Data on report timeliness not collected in 2003; time from Gram-positive rod growth to report missing for 15 isolates in 2004.
From 2004 to 2006, the years in which report timeliness data were collected, the median time from laboratory recognition of a positive culture to making a telephone report was 3·3 h. Telephone reports received within 12 h of GPR positivity steadily increased from 60% to 79% (P<0·01); reports received within 24 h increased from 76% to 91% (P<0·01).
No highly suspect cases of anthrax were identified by clinical follow-up; no isolates needed rapid transportation to the CDPH laboratory to immediately rule out anthrax. Of the 386 Bacillus spp. isolates with known haemolysis and motility, 69 (18%) were non-haemolytic and non-motile; 56 (81%) of these were forwarded to the CDPH laboratory to rule out B. anthracis by gamma-phage lysis; all were negative. There were no Clostridium spp. outbreaks identified and no individual cases attributed to contaminated allografts, injection drug use, or recent medical abortion.
Figure 2 presents the distribution of daily counts of GPR isolates for the 4-year period by collection date for isolates meeting the telephone reporting requirement of being identified within 32 h of culture inoculation. On average, 0·81 isolates were identified per day of collection, an average of one telephone report every 3 days per million residents. Significant clusters of GPR isolate growth (⩾4 reports per day) were identified on only 14 (1%) of surveillance days. Follow-up of individual reports within these clusters verified that none of these resulted from an outbreak of specific illness.
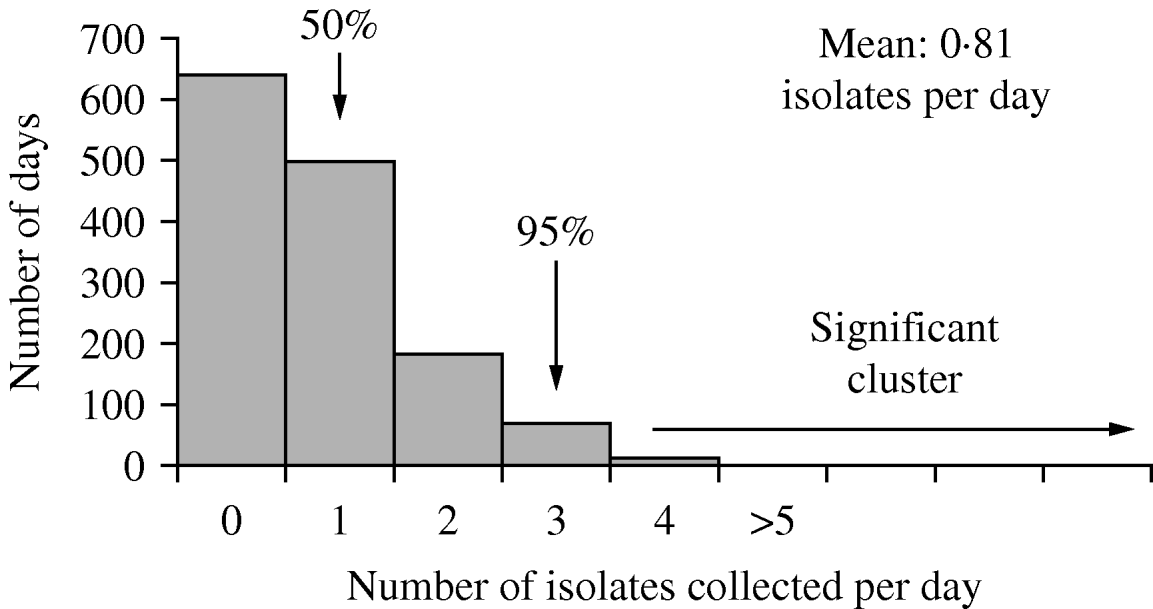
Fig. 2. Distribution of daily counts of Gram-positive rod isolates in Connecticut, 1 March to 31 December 2006.
Figure 3 shows the comparison of the investigation timeline for the case of inhalational anthrax that occurred in Pennsylvania to what might have optimally occurred in Connecticut. The Pennsylvania patient was hospitalized on 16 February 2006 with pneumonia; the next day, GPRs were detected in all four of the patient's blood-culture specimens. In Connecticut, these preliminary results would have triggered a call to CDPH by the hospital laboratory and immediate clinical follow-up by CDPH staff. The findings of GPRs in multiple blood cultures, coupled with the diagnosis of community-onset pneumonia and an occupational history of drum making, would have raised the index of suspicion for inhalational anthrax. Aliquots of the positive blood specimens would have been transported that same day to the CDPH laboratory for polymerase chain reaction (PCR) testing and culture confirmation. Preliminary PCR testing yielding a positive result for B. anthracis would have been reported to CDC and the New York City (NYC) Department of Health and Mental Hygiene (DHMH), followed by culture confirmation by the CDPH laboratory on 18 February. In Pennsylvania, where anthrax is reportable but no GPR surveillance system is in place [20], the hospital laboratory first contacted the Pennsylvania Department of Health (PDH) to report suspected B. anthracis on 20 February; PDH confirmed the finding on 21 February and subsequently reported to CDC and NYC DHMH a case of inhalational anthrax in a man who resided in New York City. Based on this, Connecticut would have probably been aware of and confirmed B. anthracis 3 days earlier than Pennsylvania, therefore initiation of control measures would have been possible 3 days earlier.
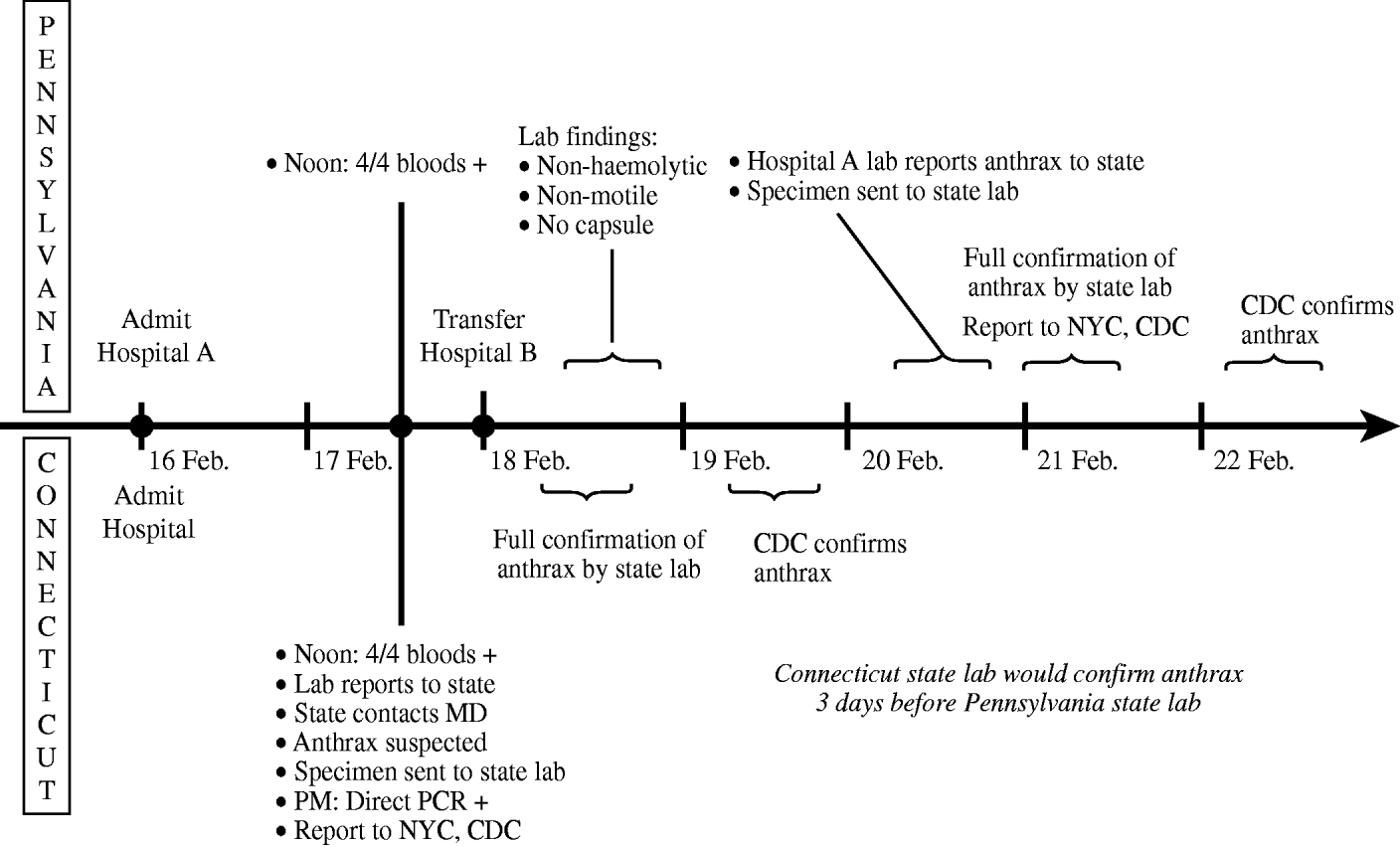
Fig. 3. Timeline of actual (Pennsylvania) vs. hypothetical (Connecticut) diagnosis of anthrax, February 2006. CDC, Centers for Disease Control and Prevention; CDPH, Connecticut Department of Public Health; PCR, polymerase chain reaction.
DISCUSSION
Increasing the sensitivity and timeliness of recognition of possible outbreaks of Category A biological terrorism-associated diseases continues to be a public health preparedness challenge. Nationally, systems have been developed based on syndromes, laboratory testing results and over-the-counter pharmacy sales data, as well as outdoor and indoor environmental sampling data [Reference Bradley12–Reference Meehan14]. Systems such as these have been criticized for a number of reasons, including their expense, lack of sensitivity and specificity and the false premise of timeliness due to the time it takes to investigate a signal from these systems, determine whether there is an outbreak and identify an aetiological agent [Reference Balter15–16, 21]. For Connecticut, these issues have been averted for anthrax with a GPR surveillance system that enables rapid recognition of a single or sentinel case of inhalational anthrax through telephone reporting and immediate investigation of all isolates suspect for anthrax with <1 full-time staff position [Reference Begier7]. As described above, this system, functioning optimally, would probably have confirmed the 2006 Pennsylvania inhalational anthrax case and enabled initiation of control efforts 3 days sooner than traditional reporting. Even in a ‘less than optimal’ scenario, where reporting is delayed or clinical assessment does not result in the specimen being transported and immediately tested, each day saved by earlier detection provides potential for the important range of previously listed public health interventions.
In the Sverdlovsk anthrax incident, the one large-scale point-source inhalational anthrax outbreak that described incubation periods, an average of 6% (4/62 total cases) of cases developed anthrax each day from day 4 to day 13 after exposure [Reference Meselson4]. We believe that the system achieves the balance of strengthening proven approaches, such as health department communication with laboratories and clinicians regarding infectious disease diagnosis, and implementing novel approaches, such as real-time laboratory reporting of initial test results. Such balance is recommended by the United States Institute of Medicine for early outbreak detection [Reference Smolinski, Hamburg and Lederberg22].
The GPR reporting system in Connecticut has proven to be sensitive, timely and sustainable, with system sensitivity and timeliness improving over the 4 years since its implementation. In addition, it is a specific system, with the potential for an aetiological diagnosis <24 h after the initial GPR signal. Annual follow-up with laboratories has been conducted to emphasize the need for complete reporting and the importance of forwarding all non-motile, non-haemolytic Bacillus spp. isolates to the CDPH laboratory to rule out B. anthracis. Despite not having had an inhalational anthrax case during this time, an all-time high of 88% of all such isolates were sent to the CDPH laboratory in 2006 for further confirmation.
The system does have limitations. While it helps in early detection of anthrax and Clostridium spp. outbreaks, it is not designed to detect other agents or outbreaks of potential public health importance. Second, while we report a successful surveillance system, as demonstrated by a low rate of reports found on laboratory audit (7% in 2006), high rates of telephone reporting (92% in 2006), and timeliness of these telephone reports (91% within 24 h), there is still potential for a single report to be missed early on. If this were an index case of anthrax, the advantages of timely detection using this system would be lost. Third, systems based on anthrax-positive laboratory test findings are of limited value when patients received antimicrobials prior to specimen collection. The three blood-culture-negative inhalational anthrax cases diagnosed in the 2001 mail attacks had all received antibiotics prior to blood cultures being obtained [Reference Jernigan2]. Fourth, this system might require one or more dedicated full-time position(s) in public health jurisdictions with much larger populations and many more clinical laboratories. Finally, we did not carry out a thorough examination of the acceptability of the system to clinical laboratories. However, given that there is only an average of one case reported statewide every day and that there are 34 clinical laboratories in Connecticut, the incremental workload for any given laboratory is very small.
In summary, with the use of laboratory audits and ongoing evaluation, CDPH has instituted and maintained a sensitive, almost complete, round-the-clock reporting system for timely recognition of possible inhalational or septic anthrax cases that reduces dependence on astute clinicians. The system is an attempt to minimize the time lapse between the earliest specific diagnostic finding and notification of the public health system. Given the potential to detect an initial case of inhalational anthrax several days earlier with Connecticut's system, public health jurisdictions at any governmental level interested in as timely as possible detection of and response to a large-scale anthrax outbreak should consider implementing a similar system where resources permit.
ACKNOWLEDGEMENTS
Many thanks to Nancy L. Barrett, who helped spearhead efforts to implement and improve this GPR surveillance system. Thanks are also due to Connecticut's laboratories and clinicians for making this system possible, to M. Zack Fraser for his work with the GPR surveillance system and laboratory audits, and the additional members of the 2003–2006 Connecticut Bioterrorism Field Epidemiology Team (Kasia Frenette, Lisa LoBianco Pippa, and Ava Nepaul).
DECLARATION OF INTEREST
None.







