Introduction
The Betacoronavirus genus includes multiple bat-associated viruses and several human pathogens, including severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle Eastern respiratory syndrome coronavirus (MERS-CoV) [Reference Woo1, Reference Drexler, Corman and Drosten2]. In 2013, a novel clade C Betacoronavirus was discovered in the Western European hedgehog (Erinaceus europaeus) in Germany [Reference Corman3]. The virus was subsequently detected in the same species in France [Reference Monchatre-Leroy4]. Characterisation of these Erinaceus coronavirus (EriCoV) nucleotide sequences revealed high nucleotide identity to MERS-CoV [Reference Corman3], the cause of an acute respiratory syndrome in humans with high case fatality rates [Reference Memish5, Reference Mohd, Al-Tawfiq and Memish6].
The structural spike protein is key to coronavirus virulence and host specificity [Reference Bosch7] and the gene that encodes it plays a pivotal role in coronavirus diversity [Reference Graham and Baric8]. Analysis of the receptor-binding region in the S protein of EriCoV revealed that it shared only 36.7% amino acid identity with that of MERS-CoV [Reference Corman3]. However, there have been no further published EriCoV whole genome sequencing attempts, and many questions remain over the origins and evolution of EriCoV [Reference Drexler, Corman and Drosten2]. Whilst there is no known threat to public health from EriCoV, further investigation of the virus is warranted to gain a fuller understanding of the impact of coronavirus infection on wildlife populations.
Bats are widely recognised and accepted as the natural reservoir hosts of a variety of coronavirus species, including SARS-CoV [Reference Li9, Reference Hu10]. Both bats and camels have been cited as possible reservoirs of MERS-CoV [Reference Mohd, Al-Tawfiq and Memish6, Reference Milne-Price, Miazgowicz and Munster11]. It is considered that bats are the likely natural reservoir of MERS-CoV [Reference Drexler, Corman and Drosten2, Reference Memish5], and that dromedaries (Camelus dromedarius), and potentially other species, act as an intermediate host for human transmission [Reference Mohd, Al-Tawfiq and Memish6, Reference Haagmans12, Reference Hemida13]. MERS-CoV-like virus sequences have only been detected in one other species, E. europaeus [Reference Corman3]. Hedgehogs are members of the order Eulipotyphla, which is closely related to Chiroptera, the order of bats [Reference Onuma14]. This close association is what initially led to hedgehogs being investigated as a potential coronavirus reservoir [Reference Corman3].
Many animal species seem to have the capacity for coronavirus infection in the absence of apparent disease, including bats [Reference Lau15], aquatic birds [Reference Chu16] and rabbits when inoculated with MERS-CoV [Reference Haagmans17]. However, dromedaries infected with MERS-CoV develop pyrexia with mild upper respiratory signs, consisting of serous to purulent nasal discharge [Reference Adney18]. Lesions in infected dromedaries are comparable to those seen in the common cold in humans, including inflammation and loss of pseudostratified epithelial cells in the upper respiratory tract. Coronaviruses that affect the gastrointestinal system, such as porcine transmissible gastroenteritis virus, typically cause similarly mild lesions, including destruction of enterocytes and villus atrophy [Reference Kim19].
Although still widespread throughout much of Western and Northern Europe, the Western European hedgehog population in Great Britain (GB) has seen a rapid decline in recent decades, with an estimated population decrease of 95% between the 1950s and 1990s [Reference Reeve20, Reference Wembridge21]. The reasons for this decline are not fully understood, and it is likely to be multi-factorial (e.g. habitat loss and fragmentation, road traffic death and predation) [Reference Wembridge21, Reference Pettett22]. The possible role of infectious disease also needs to be investigated, including the consequences of infection with newly-discovered viral agents. Here, we identify the presence of EriCoV in British hedgehogs, sequence the full genome, explore the spatial distribution of infection and investigate if there is any association of EriCoV with disease.
Methods
Sample collection and preparation
A total of 351 samples from individual hedgehogs were tested in this study. Of these, 225 were voided faecal samples collected from live hedgehogs that had been found injured, sick or abandoned juveniles and were consequently admitted to wildlife centres for treatment and rehabilitation in 2014 and 2015. A total of nine wildlife centres across GB collected and supplied samples, using the methods described by Sangster et al. [Reference Sangster23]. A fresh faecal sample was collected non-invasively shortly after admission and associated metadata, including clinical signs, signalment, location and history, were collected where available. Faecal samples were stored at 4 or −20 °C for a maximum of 1 week at rehabilitation centres prior to submission. A subsample of each sample collected by Sangster et al. [Reference Sangster23] was used in the current study. An additional four voided faecal samples were collected from the wild as part of a Greater London-based hedgehog population survey conducted in 2014 to give a total of 229 faecal samples from live animals.
The remaining 122 samples were of distal large intestinal tract contents collected from dead hedgehogs during post-mortem examination (PME) from 2011 to 2015 and comprised animals with infectious and non-infectious (e.g. trauma) causes of death. These hedgehogs had either died in wildlife centres within 48 h of admission or had been found dead by members of the public. Where possible, PMEs were performed on fresh carcases, but frozen carcases that had been archived at −20 °C were also examined. Systematic examination of external and internal body systems was performed using a standardised protocol, followed by microbiological, parasitological and histological investigations where indicated by macroscopic findings and when permitted by the state of carcase preservation [Reference Franklinos24]. The contents of the distal large intestinal tract were sampled to most closely reflect freshly voided faeces, as the latter had contained the highest EriCoV viral titre in a previous study [Reference Corman3].
Clinical data and case history
Faecal sample submission forms provided information on: admittance date, sample collection date, the location where the hedgehog was found, sex, live body weight, faecal characteristics (including colour and consistency) and reason for admittance. PME records were available for 121 of 122 hedgehogs from which samples of distal large intestinal contents were tested. These comprised data on the location where the dead hedgehog was found, the date the carcase was found, sex, age class, carcase weight, subjective assessment of body condition (based on muscle mass and body fat deposits), gross PME findings and the results of ancillary diagnostic tests.
Viral RNA extraction
RNA stabilising agent RNAlater (Ambion, Life Technologies Europe, NL) was added to, and mixed with, a pea-sized amount of each sample (faeces or distal large intestinal tract contents) in a 1.5 ml microcentrifuge tube. Each sample was then stored at either −20 or −80 °C. Faeces were homogenised and pooled (five samples per pool). Each 140 µl of total pooled sample was added to 560 µl buffer AVL-carrier RNA solution and extracted immediately following the spin protocol alongside negative extraction controls. A guanidine and column-based method (QIAamp Viral Mini Kit Qiagen, Hilden, DE) was used for RNA extraction, according to the manufacturer's protocol. RNA was then eluted in 60 µl of buffer AVE and stored at −80 °C.
Detection of EriCoV RNA
A specific oligonucleotide primer set (EriCoVR and EriCoVF, see Supplementary Table S1) was designed based on the EriCoV RNA-dependent RNA-polymerase (RdRp) gene sequence of the German virus sequence (KC545383) with OligoArchitect™ (Sigma-Aldrich®, St Louis, MO, USA) and used with the BRYT-Green based GoTaq® 1-Step real-time reverse transcription PCR (RT-PCR) system kit (Promega Corporation, Madison, WI, USA) to detect EriCoV RNA in an Applied Biosystems 7500 Fast Real-Time system. A 10−2 dilution of extracted RNA from a positive EriCoV sample (R618/14, previously confirmed through sequencing) was used as the positive control. Nuclease-free water (Promega Corporation) was used as the negative control. Following the amplification stages, melt curve analysis was performed in order to verify the specificity of the amplicons [Reference Ririe, Rasmussen and Wittwer25].
In addition, a 110 bp EriCoV ultramer oligonucleotide was used to quantify the limits of detection and assess the sensitivity of the designed assay. This was a sequence within the EriCoV primer RdRp target region, but with the removal of 10 nucleotides at the centre to differentiate it from wild-type EriCoV (see Supplementary Table S1). A 14-fold dilution series was performed and a standard curve was generated using GraphPad Prism 6 (GraphPad Software, CA, USA). To assess assay specificity, RNA of the gammacoronavirus, infectious bronchitis virus (IBV), was also included as a control.
To assess the primer dynamic range and potential RNA degradation, a dilution series of RNA extracted from faeces was tested. A ‘spiked’ dilution series was set containing 25% (5 µl in 20 µl) faecal RNA extraction from an EriCoV-negative pool and a subset of negative samples (n = 11) was tested using primers for the 18S ribosomal RNA subunit. All samples extracted from faeces produced positive results confirming the presence of amplifiable RNA (data not shown).
Real-time RT-PCR assays using the EriCoV primers were carried out for all pooled samples. Single samples from positive pools were then extracted and tested individually. A subset of amplicons from individual samples (n = 10) was also assessed by gel electrophoresis and compared to the positive control. Sanger sequencing was conducted on three amplicons with EriCoVR and EriCoVF primers using standard protocols. Amplicon sequences were manually checked then aligned against the German EriCoV RdRp gene fragment sequence retrieved from GenBank (accession number KC545385) to confirm their identity (MEGA 6.0).
Whole genome sequencing
Whole genome sequencing was performed on RNA extracted from one faecal sample collected in 2014 (R618/14) which was identified as EriCoV-positive by real-time RT-PCR. Sequencing was performed using synthesis technology (Illumina, San Diego, CA, USA). RNA was extracted as before and DNA was depleted from this sample using the on-column DNase treatment (RNeasy® plus mini kit) following the manufacturer's instructions (Qiagen); the resultant RNA was then eluted in 30 µl molecular grade water [Reference Marston26]. cDNA was synthesised from 50 ng RNA using a random cDNA synthesis system (cDNA Synthesis Kit, Merck, De), according to the manufacturers’ instructions. The cDNA was purified using Ampure XP magnetic beads (Beckman Coulter, High Wycombe, UK), 1 ng of which was processed for sequencing using the Nextera XT DNA sample preparation kit (Illumina). A sequencing library was prepared according to the manufacturers’ instructions and sequenced using an Illumina MiSeq with 2 × 150 bp paired end reads following standard Illumina protocols.
For confirmation of the insertions and deletions of the resulting sequence, seven extra sets of primers were designed (available on request). The sequences of these short amplicons were compared with the sequence derived by Illumina sequencing. The total reads (12 908 682) were mapped to a reference EriCoV sequence from Germany (GenBank accession number KC545383). A modified SAMtools/vcfutils script was used to generate an intermediate consensus sequence in which any indels relative to the original reference sequence were appropriately called. The intermediate consensus was used as the reference for subsequent iteration of mapping and consensus calling. The total number of assembled viral reads was 1 217 783 (9.43% of the total reads). Despite the low proportion of viral sequence detected within the total data set, coverage of the entire genome was obtained (average coverage depth of 4546 reads).
Post-mortem analysis and histopathological investigation
Gross PME findings were categorised into whether respiratory abnormalities (including, but not limited to, pneumonia, consolidation, haemorrhage, congestion, granuloma and high Crenosoma sp. burden) and/or gastrointestinal abnormalities (including, but not limited to, haemorrhage, inflammation, intussusception, rupture, abscess and faeces of abnormal colour or consistency) were detected. Gross PME findings were available for both organ systems from 94 of the 121 hedgehog examined post mortem and were included in these analyses.
A subset of hedgehogs (N = 9) examined postmortem that were EriCoV-positive was selected for histological examination. Selection criteria focused on carcases likely to provide the optimal state of tissue preservation from those available, prioritising tissues from carcases examined fresh or when frozen, from carcases with the least evidence of autolysis. A range of available tissue samples from each selected carcase was fixed in neutral-buffered 10% formalin and was prepared for histopathological examination and stained with haematoxylin and eosin using standard techniques. Particular attention was paid to examination of the respiratory and alimentary tracts, when available, as these are the organ systems frequently targeted by mammalian CoVs.
Statistical analysis
Associations between EriCoV-positive status, case history signalment and pathological findings were explored. Variables were first re-categorised and coded using Microsoft Excel before being exported to IBM® SPSS® version 22 for Windows (IBM®, Chicago, IL, USA) for statistical analyses. Cross-tabulation and Pearson's χ 2 tests were undertaken for the binary dependent variable (EriCoV presence or absence) against all other categorical independent variables. In addition, where sample numbers were sufficient, binary logistic regression (BLR) was performed to include consideration of continuous independent variables, such as age, and to quantify the strength of association for multiple variables simultaneously (year, region). A P-value of <0.05 was considered statistically significant. ArcGIS Pro version 11 software (Esri, Redlands, CA, USA) was used to map the distribution of hedgehogs for which location data were available.
Results
Detection of EriCoV
Of 351 samples examined, 38 (24 faecal samples and 14 distal large intestinal tract samples) were positive for EriCoV, giving a positive percentage of 10.8%. The viral copies detected in positive faecal samples ranged from 1.15 × 108 to 2.7X104 per 4 µl extracted RNA. The limit of detection for the BRYT® Green I based EriCoV primer RT qPCR assay was calculated as 2.68 × 103 copies per 1 µl of extracted RNA sample. IBV RNA at 10−2 dilution was not detected by the EriCoV-specific assay.
Whole genome sequencing
The complete genome of the hedgehog EriCoV from sample (R618/14, Genbank accession number: MK679660) is 30 173 nucleotides (nt) in length, which is comparable to those sequenced from Germany: KC545383 and KC545386 (30 148 and 30 175 nt, respectively). Complete genome comparison with open reading frames (ORFs) from the German EriCoV (KC545383) gave identities ranging from 79% (ORF3b) to 98% (ORF6, encoding the envelope protein). The genes encoding the spike protein of the British and German EriCoV viruses shared 93% identity (Table 1).
Table 1. Comparison of open reading frame (ORF) lengths of Erinaceus coronavirus from Great Britain (GB) and German hedgehogs (Erinaceus europaeus)
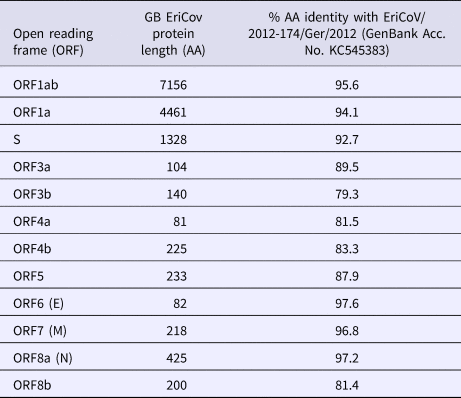
AA, amino acid
Geospatial analysis
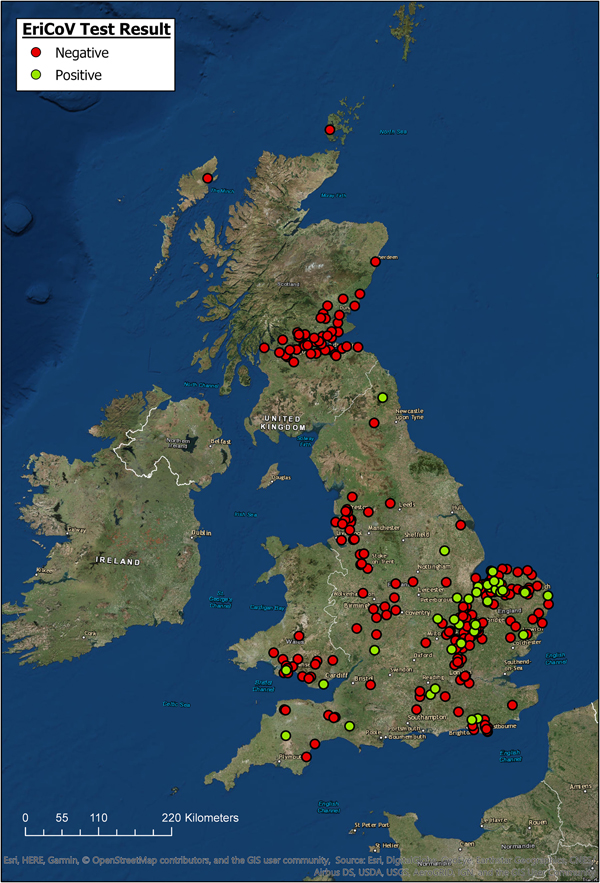
The locations for all hedgehogs with available data (n = 344) are displayed in Figure 1, along with their EriCoV status. Hedgehogs sampled for the study covered a wide area across GB, from Devon in South West England to Northern Scotland. Hedgehogs positive for EriCoV were found in Southern England, East of England, North East England and Wales. We found no evidence of EriCoV in hedgehogs from Scotland, even though 17.4% (n = 61) of sampled hedgehogs were submitted from this country. There was a bias of submissions from the East of England, accounting for 44.4% (n = 123) of the total, with Norfolk alone contributing 23.1% (n = 64) of the hedgehog samples examined, which is explained by a single, large participating wildlife centre in this region. Although all regions of GB were represented, some appeared to be under-represented; for example, only 1.1% (n = 3) of submissions came from North East England.
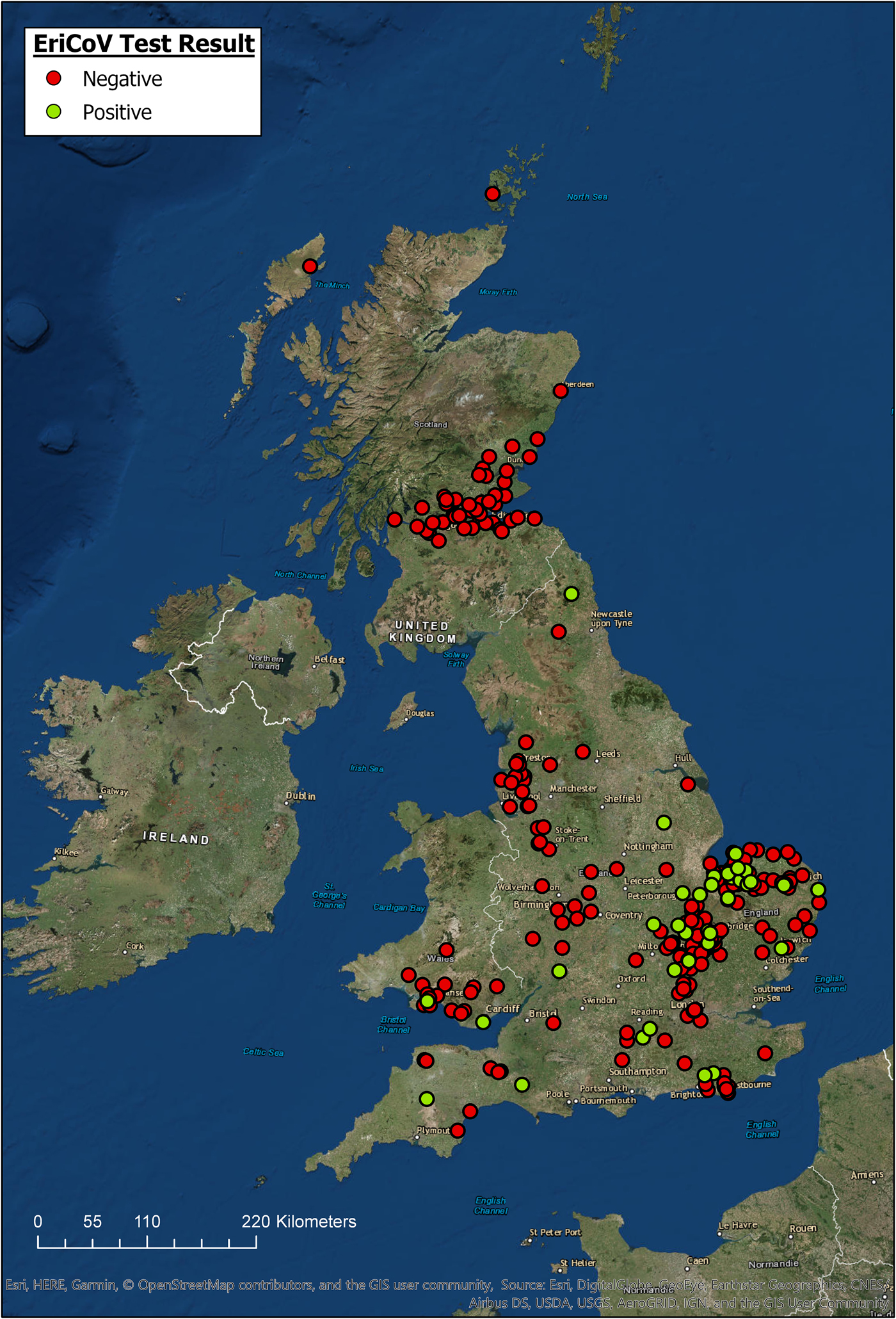
Fig. 1. Geographic origin of hedgehog samples and EriCoV testing result (ArcGIS).
Clinical and pathological associations with EriCoV detection
The majority of hedgehog samples were submitted in 2014 and 2015. The proportion positive in 2015 was higher (28/144, 19%) than in 2014 (10/182, 5%) but there were too few submissions from other years to determine if there was an association with year of sampling. There was a higher number of juvenile hedgehogs included in the study in 2015 (n = 68) than 2014 (n = 24), but the proportion of juveniles positive in 2015 (19.1%) was comparable to the proportion of adults positive (20.3%).
As coronaviruses are associated with diarrhoea in other species, we tested for the possibility of an association between EriCoV status and abnormal faeces. The proportion of samples from hedgehogs with reportedly normal faecal colour or consistency, which tested positive for EriCoV (14/102), was not significantly different from the proportion of all faecal samples with abnormal consistency and/or abnormal colour (8/115) (P = 0.099). However, a significant association was observed between EriCoV-positive hedgehogs and those with green faeces (P = 0.004). A significant association was also observed between EriCoV-positive hedgehogs and those with yellow faeces (P = 0.034). Colour variation within a single sample of hedgehog faeces was not uncommon: 42.6% (n = 20) of hedgehogs with green coloured faeces were also reported to have faeces of brown, yellow or black colouration. No other statistically significant association was shown across the other independent variables, including body condition (Table 2). A BLR model testing all but the non-mutually exclusive variables also showed no statistical significance.
Table 2. Associations with EriCoV infection
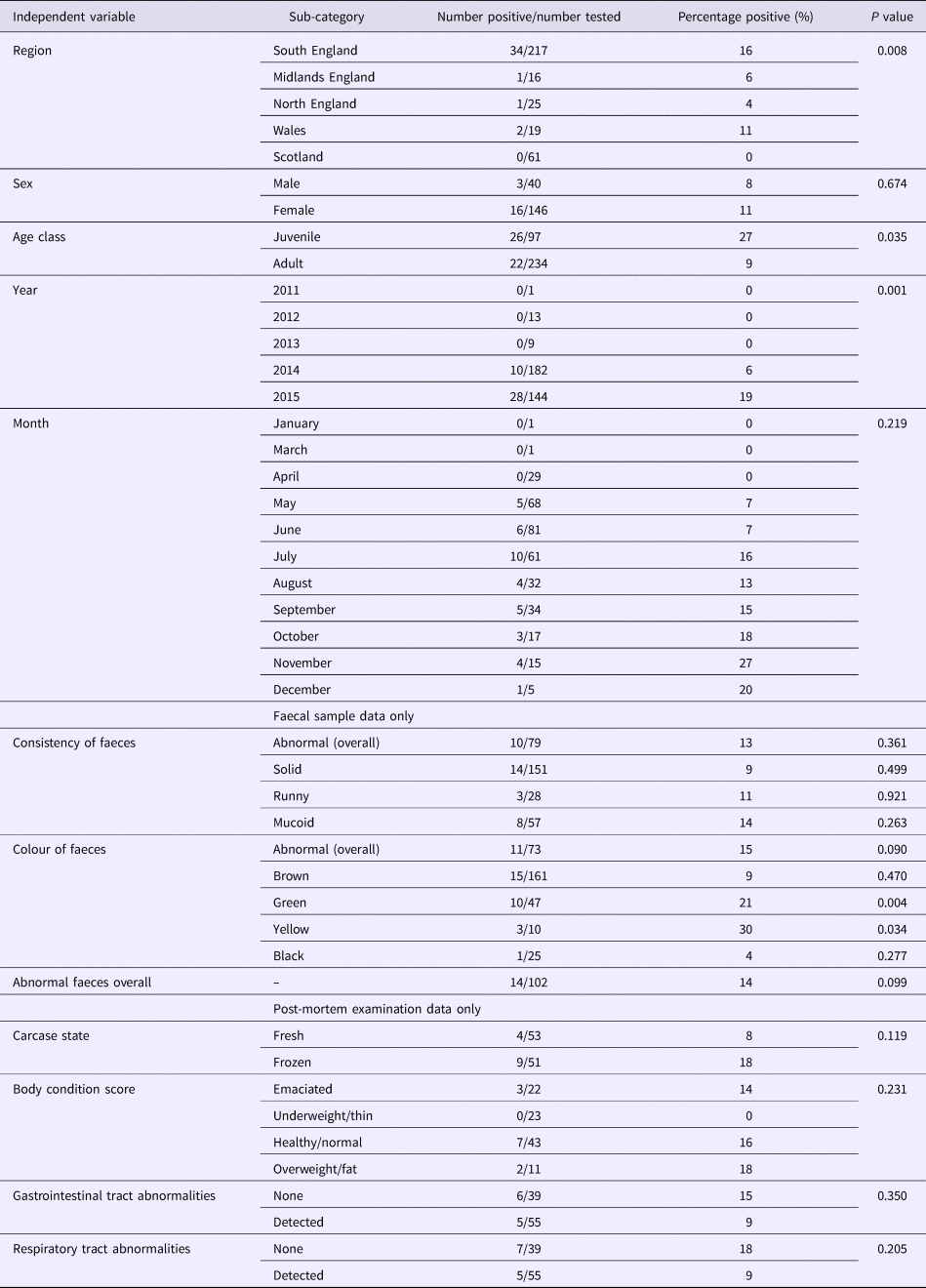
A P-value of <0.05 was considered statistically significant.
The proportion of hedgehogs with respiratory abnormalities reported at PME that tested positive for Eri-CoV (5/55) was not significantly different from the proportion of those without respiratory abnormalities that tested positive (7/39) (P = 0.205). The proportion of hedgehogs with gastrointestinal tract (GIT) abnormalities that tested positive (5/55) was not significantly different from those that had no GIT abnormalities (6/39) (P = 0.35).
A statistically significant association was shown between EriCoV status and geographical region of origin (P = 0.008) (Table 2). The highest proportion of EriCoV-positive hedgehog samples were submitted from the South of England (34/217, 16%); however, BLR showed no significant association (P = 0.678) between EriCoV infection status and wider region when other factors including age and year were included.
Histopathology
Histopathological examination was conducted on available tissues from nine of the EriCoV-positive hedgehogs in this study that were examined post-mortem: results are presented with the signalment, macroscopic findings and other ancillary diagnostic tests for interpretation in Supplementary Table S2. Interpretation was complicated by the state of tissue preservation in the majority of cases, limiting the ability to appraise evidence of either a host inflammatory response or the presence of mild abnormalities (e.g. loss of pseudostratified epithelial cells in the respiratory tract or intestinal villus atrophy, as has been reported with some coronavirus infections in other species [Reference Perlman27]). Examination of the respiratory and gastro-intestinal tract identified frequent metazoan parasite infections of variable severity, sometimes in combination with bronchopneumonia. A range of other significant findings was detected, supported by ancillary testing, including trauma and yersiniosis.
Discussion
We detected EriCoV, a novel clade C Betacoronavirus, in wild Western European hedgehogs in GB. This virus had previously only been reported from Germany and France; in both instances from this same species. Our results, therefore, indicate that EriCoV is widespread in this European wild mammal. Full genome sequence of EriCoV from GB showed 94% identity with a EriCoV previously detected in Germany, and 78% identity with the virus responsible for MERS [Reference Corman3].
There was no evidence to indicate that EriCoV was causing clinical disease in hedgehogs. The lack of association with detection of EriCoV and abnormal faecal consistency, poor body condition (i.e. thin or emaciated), respiratory and digestive tract abnormalities suggests EriCoV is unlikely to be a primary pathogen in this species. This concurs with studies reporting natural CoV infections in wild animal species, including bats [Reference Lau15] and aquatic birds [Reference Chu16], which do not manifest as clinical disease. This is also a common feature of a reservoir or maintenance host [Reference Haydon28, Reference Viana29]. Coronavirus infections of man and other animals are known to predispose to secondary infections, for example, a study recently isolated feline coronavirus and several other enteropathogens from cats with diarrhoea [Reference Paris30], thus making any association difficult to detect should it occur with EriCoV infection in the hedgehog. In the current study, histopathological examination was complicated by concurrent parasitic and bacterial infections in several cases and limited by freeze–thaw artefact and the state of tissue preservation, therefore microscopic lesions caused by viral infection could have been missed or obscured. Experimental infection studies and the development of in situ hybridisation to localise EriCoV in tissues would be worthwhile in the future to further elucidate the clinical significance of EriCoV infection.
Despite the lack of association between EriCoV and faecal abnormalities overall, there was an apparent association between EriCoV status and green or yellow faeces. Green faeces is not an unusual finding in hedgehogs, and can often be associated with Crenosoma sp., Isospora or Salmonella sp. infection [Reference Bunnell31]. Many of the hedgehogs reported to have green faeces also had faeces of another colour, suggesting that faecal colour change is not unusual. The colour change or the reporting thereof may not be consistent and therefore this result should be interpreted with caution. However, the possibility of a causative relationship between EriCoV infection and abnormal faeces colour cannot be excluded.
Significantly stronger amplification of CoV RNA has previously been described in juvenile and lactating female Myotis sp. bats [Reference Drexler32], and younger age has been shown to be associated with increased coronavirus shedding across various bat species [Reference Mendenhall33, Reference Osborne34]. Although no evidence for such an association for EriCoV in the hedgehog was found in the current study, juveniles accounted for only 27.6% of the hedgehogs sampled. Examination of a larger sample size of juvenile animals would be worthwhile to further explore the possibility of age-structured prevalence.
Hedgehogs positive for EriCoV were detected over a wide region of England and Wales, but not from Scotland even though 17.4% of all samples tested were submitted from this country (Fig. 1). Inferring population prevalence from these data is inappropriate due to the sampling strategy, relying on convenience sampling and the catchment area of rehabilitation centres. There is apparent sampling bias since 47% of all hedgehog samples were from the East of England. Demonstrating regional differences in population prevalence and absence of infection would require a much larger, random sample of the population, and information regarding the population structure of hedgehogs in GB.
Annual variation in the proportion of EriCoV-positive samples was found. The proportion of hedgehogs sampled in 2015 that tested positive for EriCoV was higher than that in 2014; a difference that was consistent irrespective of sample type or hedgehog age class (Table 2). This temporal variability in detection rate is consistent with previous multi-year studies in bats where significant annual variation in coronavirus detection rate was reported [Reference Osborne34]. Seasonal variation in contact rates, environmental factors and fluctuating levels of CoV immunity in the population have all been postulated as causes for such fluctuations [Reference Osborne34]. Proving viral persistence and establishing critical community size is challenging for wildlife populations [Reference Viana29]. Whilst there is now a wealth of studies investigating bat CoVs, research is lacking for other wildlife CoVs. There is evidence that the gregarious roosting habits of some species of bat facilitate CoV transmission [Reference Wacharapluesadee35], but the transmission dynamics of EriCoV in the more-solitary hedgehog [Reference Reeve20] are likely to be very different. Establishing whether infection is endemic in British hedgehogs will require longitudinal sampling, augmented by age-specific serology [Reference Gilbert36].
Overall, the percentage of EriCoV-positive hedgehogs detected, at 10.8%, was lower than that reported previously, where 58.9% (148/248) and 50% (37/74) of specimens tested positive for EriCoV in Germany and France, respectively [Reference Corman3, Reference Monchatre-Leroy4]. This difference could be due to assay sensitivity or sample degradation. Although the limit of detection of the previously used assays was not given, average copy numbers in the faecal samples were well over the sensitivity threshold of the assay used here. All three studies included hedgehogs in wildlife centres. In the German study, no details were given for the timing of sampling. In the current and French studies, however, faecal samples were taken soon after admission (typically within 48 h) in order to reduce the possibility of nosocomial transmission. Dog kennels and shelters are recognised as important environments for maintaining enteric canine CoVs, with increased length of stay associated with increased probability of CoV infection [Reference Stavisky37]. However, nosocomial transmission seems to be an unlikely cause of the differences in sample percentage of positive samples in this study. Another potential factor is that the relatively high percentage of positive samples in the French and German studies could be simply due to a much higher prevalence of EriCoV in the mainland Europe hedgehog population: the recruited wildlife centres (from which the hedgehogs were sampled) were all located in North and North Western France, with one located close to the border with Germany [Reference Monchatre-Leroy4]. More studies are required to determine this.
Hedgehogs in Northern Europe hibernate for 2–5 months of the year [Reference Reeve20]. EriCoV was detected in both 2014 and 2015, suggesting that the virus overwinters in the hedgehog population, but the method of viral persistence is not known. In the current study, no association was detected with the month of sampling but to explore this further, longitudinal sampling of individual hedgehogs could be conducted, including from animals before and after hibernation, to determine if there is persistence of infection in individuals.
Deriving full genome sequence is an important step in characterising this virus to assess relationships with other CoVs, including pathogens of domestic animals and man. Overall identity to the previous coronavirus detected in hedgehogs from mainland Europe was high (94%), but lower than the identity between two German EriCoV isolates (97%) [Reference Corman3]. Previous studies have demonstrated that EriCoV shares a common ancestor with MERS-CoV, and bat coronaviruses HKU5 and HKU4, raising the possibility of a lineage that has been positively selected for and which has evolved with the potential for cross-species transmission. Host tropism in the coronaviruses is largely determined by the spike protein and its ability to bind to host cell receptors. Both the German and the GB EriCoVs have <40% amino acid similarity with MERS-CoV at the crucial receptor-binding domain on the spike protein, suggesting that binding to the human receptor (dipeptidyl peptidase 4) is unlikely. Virus culture was not attempted in this study, but was previously unsuccessful and this might be due to the cell tropism of the virus [Reference Corman3].
The detection of EriCoV in British hedgehogs is a notable finding, and the first report outside mainland Europe. Wildlife disease research in GB has greatly increased in recent years, particularly for popular wildlife species such as the hedgehog, where citizen science offers a cost-effective approach to achieving national disease surveillance [Reference Lawson, Petrovan and Cunningham38]. It is important that this continues if we are to gain a deeper understanding of endemic and emerging pathogens of wildlife and their impact on wildlife, livestock and human populations.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268819000207.
Author ORCIDs
D. L. Horton, 0000-0002-9126-2756
Acknowledgements
We thank Lydia Franklinos, Tim Hopkins, Lucy Sangster and Shaheed Macgregor (ZSL) for assistance with sample collection; members of the public and the network of participating wildlife centres for submitting hedgehog faecal samples from casualty hedgehogs for testing (East Sussex Wildlife Rescue and Ambulance Service, Gower Bird Hospital, RSPCA East Winch, RSPCA Stapeley Grange, RSPCA West Hatch, Shepreth Hedgehog Hospital, The Scottish Society for the Prevention of Cruelty to Animals and Vale Wildlife Hospital and Rehabilitation Centre).
Financial support
This research was undertaken as part of the Garden Wildlife Health (GWH) project (www.gardenwildlifehealth.org). Funding for the GWH was provided by the UK Department for the Environment Food and Rural Affairs, Scottish and Welsh Government through the Animal Plant & Health Agency's Diseases of Wildlife Scheme Scanning Surveillance Programme (Project ED1600) and SV3045, the Esmée Fairbairn Foundation and the Universities Federation for Animal Welfare. Work at the University of Surrey was undertaken as part of a Master of Science degree in Veterinary Microbiology.