Acute gastroenteritis (AGE) is a major cause of morbidity and mortality worldwide. It is estimated that 1·5–2·5 million deaths and between 3 and 5 billion cases occur annually due to gastroenteritis in children aged <5 years. In older children, adolescents and adults, a low mortality and high morbidity with ~2·8 billion episodes per year have been reported [Reference Fischer Walker and Black1]. The majority of gastroenteritis episodes in children are due to different viral agents mainly rotavirus, norovirus, astrovirus and adenovirus, among which rotavirus infections are most predominant. Rotavirus, a member of the family Reoviridae, is classified into eight groups, A–H, based on the antigenicity of the inner capsid protein VP6. In humans, infection with group A rotavirus (GAR), B (GBR), C (GCR) and H (GHR) have been detected to date. Globally, GAR is the leading cause of AGE in children and infections due to GBR and GHR are detected at low levels [Reference Lahon2, Reference Matthijnssens3].
GCR infections have been reported in both sporadic and outbreak cases of gastroenteritis worldwide [Reference Luchs4, Reference Kumazaki and Usuku5]. However, its epidemiology and ecology remains unclear. Studies indicating low prevalence of antibodies against GCR in urban and high prevalence in rural populations as well as evidence of cross-species transmission highlight the role of GCR as an emerging zoonotic infection in humans, which should be constantly monitored in the future [Reference Gomara6, Reference Gabbay7]. In India, the role of GCR in AGE cases has not been reported until now, and needs to be explored when mass vaccination against GAR is being implemented. The aim of the present study was to screen the cases of AGE for the occurrence of GCR and to study its circulation pattern in western India.
The National Institute of Virology, Pune (India) is engaged in investigations of gastroenteritis outbreaks occurring mainly in western India. Faecal specimens collected from these outbreaks during the period 2006–2014, which were available in adequate quantities for testing were included in the study (Table 1). A total of 253 retrospective faecal specimens collected from AGE outbreaks in rural [Sholapur, March 2010 (n = 118); Sholapur, August 2011 (n = 42); Miraj, December 2014 (n = 33)] and urban [Mumbai, October 2006 (n = 60)] regions of Maharashtra state, western India were included in the study (Table 1). The faecal specimens collected from sporadic AGE cases (n = 147) admitted to hospitals in Pune (n = 114) and Aurangabad (n = 33), cities of Maharashtra state, during 2011–2013 were also included in the study. The median age of gastroenteritis patients was 18 years (range 22 days to 86 years). The male:female ratio of AGE patients was 1·04:1 (male 197, female 189). The specimens were collected within 48 h of hospitalization. All patients were examined for fever, number of episodes and duration of vomiting and diarrhoea, extent of dehydration and treatment. Severity of the disease was assessed by using a 0- to 20-point score and based on sum of the points, the disease was described as mild, moderate, severe and very severe using the Ruuska & Vesikari method [Reference Ruuska and Vesikari8]. Informed consent was obtained from the parents/patients prior to collection of specimens. The faecal specimens were tested previously for the presence of different bacterial (enterotoxigenic Escherichia coli, Shigella, Vibrio cholerae, Salmonella, Klebsiella) and viral agents (GAR, GBR, norovirus, adenovirus, astrovirus, Aichivirus, enterovirus) [9; Joshi et al., unpublished observations].
Table 1. Details of patients in outbreaks of acute gastroenteritis occurring in western India during 2006–2014
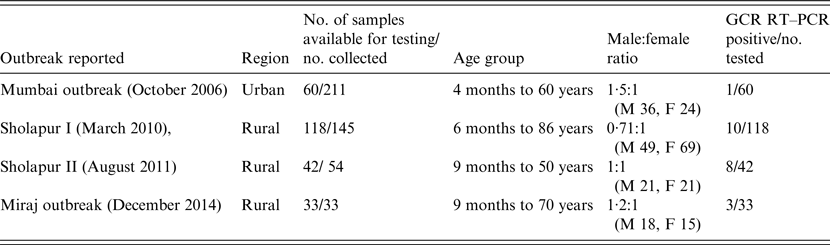
M, Male (n); F, female (n); GCR, group C rotavirus.
GCR RNA was detected by using the partial VP6 gene-based RT–PCR assay using modified C4 primer (reverse) reported by Gouvea et al. [Reference Gouvea10], MJR1: 5’-AGCCACATAGTTCACATTTCATC-3’ (1361–1339) and newly designed forward primer MJF1: 5’-ACAATWGAYATGATTAGACCAGC-3’ (891–913). Briefly, RNA was extracted from freshly prepared 30% faecal suspensions using TRIzol LS reagent (Invitrogen, USA) according to the manufacturer's protocol. The SuperScript® III One-Step RT-PCR system with Platinum® Taq DNA Polymerase kit (Invitrogen, USA) was used for both cDNA synthesis and PCR amplification in a single tube. RNA was denatured at 97 °C for 5 min and was rapidly chilled on ice for 5 min. The reaction was performed with an initial reverse transcription step at 45 °C for 30 min followed by 45 cycles of amplification (30 s at 94 °C, 30 s at 50 °C and 1 min at 68 °C) and a final extension of 5 min at 68 °C in a thermal cycler. All the PCR products were electrophoresed in 2% agarose gel containing ethidium bromide (0·5%) and visualized under a UV transilluminator. PCR amplicons were excised from the gel for purification (QIAquick, Qiagen, Germany) and cycle sequencing was conducted using a Big Dye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, USA) and an ABI 3130XL genetic analyzer (Applied Biosystems). Nucleotide sequence identity was determined through BLAST (www.ncbi.nlm.nih.gov/blast) and phylogenetic analysis was performed using MEGA 6 software [Reference Tamura11]. The phylogenetic tree was generated with a neighbour-joining algorithm and the Kimura two-parameter distance model. The reliability of the phylogenetic tree was tested by applying the bootstrap test with 1000 bootstrap replications. The nucleotide sequences of the strains examined in the study have been deposited in GenBank under the accession numbers KT900217–KT900236 and KX110051–KX110053.
Ethical approval for the study was obtained from the Institutional Human Ethical Committee of the National Institute of Virology, Pune, India.
Of the 253 and 147 faecal samples tested from outbreak and sporadic cases of AGE, 22 (8·6%) and 1 (0·7%), respectively, showed the presence of GCR RNA. The distribution of GCR positivity was 8·4% (10/118), 19·0% (8/42), 9·0% (3/33) and 1·6% (1/60) in Sholapur 2010, Sholapur 2011, Miraj 2014 and Mumbai 2006 outbreaks, respectively. In GCR-positive specimens collected from different outbreaks (n = 22), 21 were from rural regions, and 18 of these were from outbreaks occurring during March and August. Clinical severity score of the patients with GCR infection (n = 23) indicated severe disease in the majority (70%) and moderate disease in the minority (30%) of the patients (Table 2). The age distribution analysis indicated occurrence of GCR infections in patients between 9 months and 86 years with 52·1% of the cases in children (<10 years). The male:female ratio in GCR-positive patients was 2·3:1 (male 16, female 7).
Table 2. Group C rotaviruses detected as sole and mixed infections with other viral and bacterial agents in hospitalized diarrhoeic cases in western India during 2006–2014
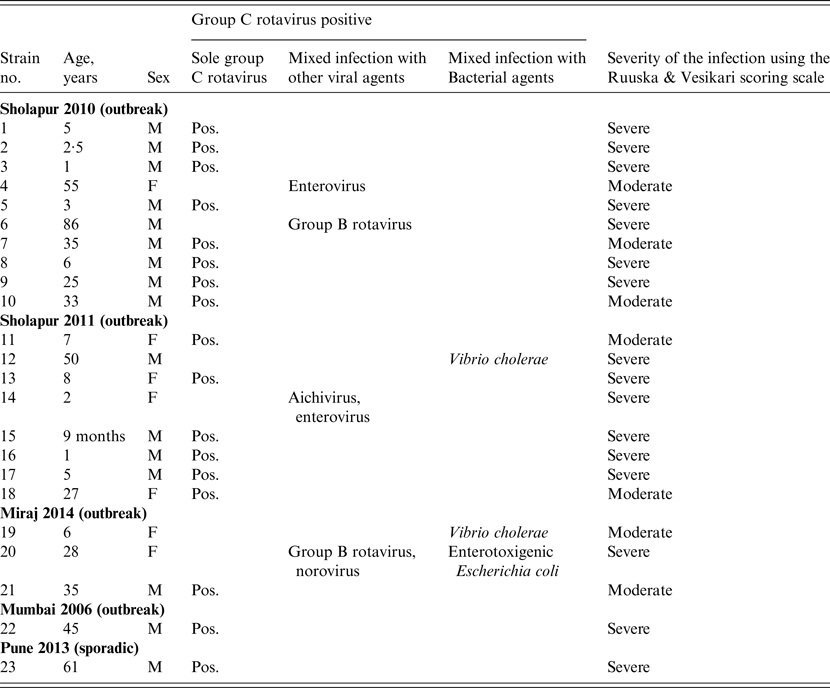
M, Male; F, female; Pos., positive.
Of the GCR-positive clinical specimens (n = 23), 17 (73·9%) were solely GCR positive while the remaining six showed mixed infection with other viral and/or bacterial agents (Table 2). GCR-associated single bacterial or viral mixed infections were detected with Vibrio cholerae (n = 2), GBR (n = 1) and enterovirus (n = 1). Mixed infections with three or four different agents were detected in one specimen each (Table 2).
Phylogenetic analysis of nucleotide sequences classified all GCR strains of the present study in to genotype I2 of the VP6 gene (Fig. 1). The study strains isolated from the outbreaks of rural regions (Sholapur 2010, Sholapur 2011, Miraj 2014) were closer to Indian strain (AY786571) and strains isolated from sporadic (Pune) and outbreak (Mumbai 2006) cases, one each from urban regions and a few outbreak cases from rural regions (Sholapur 2011, Miraj 2014), were closer to Bangladesh strain (AY754827) (Fig. 1). Nucleotide sequence identity in the 23 study strains was between 96·4% and 100%. The GCR sequences of I2 genotype randomly selected from different countries of the world, including the study strains, showed 95·1–100% nucleotide identity. The study strains showed nucleotide sequence identity in the range of 82·1–83·0% with porcine strains (I1 and I4–I7 genotypes) and 79·4% with a bovine strain (I3 genotype).

Fig. 1. Phylogenetic tree constructed based on the partial nucleotide sequences of the VP6 gene (372 bp) of group C rotavirus strains. The strains of the present study are indicted by bold font. The reference strains are indicated by accession numbers followed by the country name and year. Scale indicates genetic distances.
The present study suggests the presence of GCR strains of I2 genotype in 8·6% and 0·7% of outbreak and sporadic gastroenteritis patients, respectively. Molecular epidemiological studies available from different parts of the world indicate that GCR detection rates range between 0·3 and 23·7% [Reference Luchs4, Reference Gabbay7, Reference Phan12]. In the absence of molecular epidemiological data from India, atypical rotaviruses serologically related to GCR were detected by electrophoretic migration patterns of double-stranded RNA in 0·43% of children aged <3 years with gastroenteritis in South India [Reference Brown13]. The circulation of GCR in the southern Indian population has also been shown previously by a 25·3% seroprevalence rate [Reference Mukhopadhya14].
Among the epidemiological features, GCR-infected male patients’ outnumbered female patients (2·3:1) and infections were observed in patients of all age groups. Clinical specimens analysed from outbreaks reported from rural regions showed higher GCR positivity (10·9%) compared to outbreaks in urban regions (1·6%) and were in concordance with an earlier study indicating increased prevalence of GCR in rural regions due to the close proximity of people with animals [Reference Gabbay7]. The GCR strains circulating among diarrhoeic and non-diarrhoeic porcine and bovine animals in the study region should be studied in future in order to establish the zoonotic origin.
The study has limitations on account of the analysis being restricted to smaller numbers of faecal specimens available from sporadic (urban) and outbreak (urban and rural) cases of gastroenteritis in Maharashtra, western India. Moreover, the faecal specimens of sporadic cases collected from the rural areas and during the same period were not available for GCR screening and hence the exact role of GCR remains unknown for the region where circulation of GCR in outbreak cases has been observed. Apart from this, the primers used for the detection of GCR were specific for human and bovine GCR strains and therefore the possibility of circulation of porcine GCR strains in the study population is unexplored and warrants further study. The carriage of porcine GCR strains in diarrhoeic and non-diarrhoeic children has been reported earlier [Reference Gabbay7].
This study highlights the occurrence of GCR in outbreak cases of gastroenteritis for the first time in India. Even though the majority of GCR-positive cases were observed in rural regions, a larger prospective surveillance study is essential to reveal the exact role of GCR as well as the period of its peak activity in gastroenteritis cases in rural and urban regions of India.
ACKNOWLEDGEMENTS
We are grateful to Dr D. T. Mourya, Director, National Institute of Virology for his constant support during the study. We also acknowledge the cooperation extended by Dr S. D. Chitambar, Dr M. S. Chaddha and Dr K. S. Lole. Thanks are due to Mr Sanjay Tikute and Mr Prabhakar Jadhav for collection of specimens, Mr Atul Walimbe for statistical and phylogenetic analysis and Mr Manohar Shinde for technical assistance. The funds for the project were provided by the National Institute of Virology, Indian Council of Medical Research, Ministry of Health and Family Welfare, Government of India.
DECLARATION OF INTEREST
None.





