INTRODUCTION
Scrapie is a fatal transmissible spongiform encephalopathy (TSE) of sheep and goats. Genetic polymorphisms within the host-encoded prion protein (PrP) gene influence the risk of infection with the scrapie agent, and the age at onset of clinical disease [Reference Hunter, Piper and Ruvinsky1–Reference Tongue, Wilesmith and Cook5]. Five alleles of the PrP gene are commonly found in sheep; defined in terms of amino acids encoded at codons 136, 154 and 171 and, broadly in order of increasing risk of the classical form of the disease, these are often written as ARR, AHQ, ARH, ARQ and VRQ. These five polymorphisms form the focus of large-scale selective breeding programmes for sheep, such as the National Scrapie Plan (NSP) for Great Britain (GB) [6, 7], and wider European Union legislation applied to all member states. These selective breeding programmes, which aim to control classical scrapie, assume that the risk of classical scrapie in an individual sheep following exposure to the scrapie agent is determined predominantly by its PrP genotype, with little additional effect of other factors, such as sheep breed. This is despite the fact that there are very different incidences of classical scrapie within different sheep breeds in GB [Reference del Rio Vilas8–Reference McLean10]. The principal aim of this paper is to ask whether sheep breed plays an important role in the occurrence of classical scrapie.
We consider three routes by which sheep breed could influence scrapie occurrence. First, different breeds could harbour distinct scrapie strains, which may differentially target specific PrP genotypes. Second, breed could modulate the risk of scrapie in sheep of a specific PrP genotype; in other words, a high-risk genotype in one breed may be associated with a lower risk in another. Third, several surveys [Reference McIntyre9–Reference Gubbins11] have identified sheep breed as a significant flock-level risk factor for scrapie; in other words, some breeds have a higher proportion of affected flocks than others, after taking account of other, known risk factors. We consider whether breed-level PrP genotype frequencies, which are known to vary [Reference Eglin12], are a possible cause of the differences in scrapie occurrence across breeds.
Clear evidence for an effect of scrapie strain is from experimental studies showing that the SSBP/1 and CH1641 scrapie strains target different genotypes of Cheviot sheep [Reference Foster13–Reference Hunter15]. Field evidence for a strain effect is infection prevalences of classical and atypical scrapie in sheep sampled throughout Europe [Reference Fediaevsky16]. Surveys suggest that atypical scrapie targets several of the genotypes conventionally thought of as being at low risk of classical disease [Reference Baylis and McIntyre17, 18].
Published reports are consistent with the existence of innate breed differences in the genotype-specific risk of scrapie. For example, Suffolk sheep experience scrapie in the ARQ/ARQ genotype [Reference O'Rourke, Melco and Mickelson19, Reference Westaway20] while sheep of this same genotype in a Cheviot flock seldom succumb to disease [Reference Hunter15]. Within this Cheviot flock, the risk of scrapie in both ARQ/ARQ and ARR/VRQ sheep is approximately zero [Reference Hunter15], whereas in a flock of Romanov sheep, the risk of disease in ARQ/ARQ and ARR/VRQ animals, relative to high-risk ARQ/VRQ animals, is 50% and 0% respectively [Reference Elsen21], and in a flock of Texel sheep it is 5% and 24%, respectively [Reference Baylis22]. However, as flocks of the same breed are usually found within the same geographic region and frequently trade with each other, it is possible that distinct scrapie strains may circulate within them; if the scrapie strains differ in the genotypes they target, breed-level differences may result which are attributable to scrapie strain rather than breed per se.
To summarize, it is established that distinct scrapie strains target specific PrP genotypes but it is unknown whether the breed variability is attributable to the scrapie strains circulating within breeds, or to innate differences between the sheep breeds themselves. Here, we first verify that there are statistically significant differences between breeds in the risk of scrapie in sheep of different PrP genotypes; whether attributable to scrapie strain or sheep breed. Second, we examine the patterns of the PrP genotypes of scrapie cases in different flocks for evidence of predominant effects of scrapie strain or sheep breed. We consider the hypothesis that if sheep breed has an overwhelming effect on the PrP genotype of affected cases, then for affected flocks comprising diverse breeds, the breeds will have cases in different PrP genotypes; and for breeds with multiple affected flocks, the cases will occur in the same PrP genotypes across all flocks. Last, we investigate whether PrP genotype frequencies determine a breed's scrapie incidence.
METHODS
We undertook the following studies. (i) We estimated the risk of scrapie in specific PrP genotypes for a number of different sheep breeds. (ii) We searched for evidence that the PrP genotypes of scrapie-affected sheep do or do not differ between flocks of the same breed by undertaking two studies: (a) multiple breeds within a flock – when several different sheep breeds are combined in the same affected flock, does scrapie occur in the same genotypes regardless of breed? (b) multiple flocks within a breed – when there are many affected flocks of a specific breed, does scrapie occur in the same genotypes regardless of flock? (iii) We examined the relationship between the PrP genotype profile of a breed and the incidence of scrapie in that breed.
Datasets used
The Scrapie Notification Database (SND) held at the Veterinary Laboratories Agency (VLA) provided information on about 3500 cases of scrapie between 1 July 1998 and 31 December 2005. The 2002 anonymous postal survey of scrapie in GB [Reference McIntyre9, Reference Sivam23, Reference Sivam24] provided information on the proportion of affected flocks and flock sizes within breeds. The NSP provided breed-level data on the PrP genotypes of sheep aged >1 year. Surveys of the breeding structure of the British sheep industry in 1996 and 2003 provided data on the demography of British sheep including the flock sizes of certain breeds [Reference Pollott25, Reference Pollott and Stone26].
Statistical analyses
Risk of scrapie for different genotypes within certain breeds
The risk of scrapie for each PrP genotype was estimated for breeds with >20 cases of scrapie in SND. The measure of risk used was the estimated number of reported cases per annum per million sheep (RCAM; see [Reference Baylis3]) in breed b and genotype g, which is given by
where C bg is the number of reported cases in breed b, of genotype g (between January 2002 and December 2005, inclusive; from SND), f is a correction factor for the proportion of cases of unknown genotype (the total number of cases divided by the total number of cases of known genotype of breed, b; from SND), p bg is the proportion of animals of breed b, of genotype g (from NSP), N b is the number of sheep of breed b in GB (from [Reference Pollott and Stone26]), and T is the time period (in years) over which cases were reported. The Wilson Score Interval was used to compute 95% confidence intervals for each RCAM [Reference Agresti27]. Generalized linear models (GLMs) with binomial errors and a logit link function were used to assess differences in the RCAMs between breeds for the ARQ/ARQ, ARR/VRQ, ARQ/VRQ and VRQ/VRQ genotypes.
Flock-level variation in the PrP genotypes of cases
Cases of scrapie confirmed in SND from 1998 to 2005 were used to investigate whether or not the PrP genotypes of scrapie-affected sheep differ between flocks of the same breed. For the multiple-breed within-flock study, we examined the PrP genotypes of cases for all single flocks that had confirmed scrapie in at least three distinct breeds. For the multiple-flock within-breed study, we examined the PrP genotypes of cases for all breeds for which there were at least five reported cases in more than one flock. To preserve the anonymity of flocks affected by disease, they were alphabetically or numerically coded. As denominator data were not available for any of the flocks, a descriptive rather than an analytical approach was utilized.
Association between PrP genotype frequencies and the incidence of scrapie in different sheep breeds
To assess the effects of a theoretical risk of scrapie, computed from the PrP genotype frequency of a breed upon the observed incidence of scrapie in that breed, GLMs were employed, with logit link functions and quasi-binomial errors, which correct for over-dispersion in the data. Flock size, a risk factor associated with the acquisition of scrapie [Reference McIntyre9, Reference McLean10] for which there is variation amongst breeds, was included in the model as a covariate, log-transformed to normalize the data. All models were constructed using backward stepwise deletion of non-significant terms (P>0·05), from a maximal model which included an interaction between the estimated risk of scrapie in breeds and flock size.
The number of flocks which reported scrapie was used to estimate the incidence of scrapie in each breed. We considered three estimates of the proportion of flocks affected by scrapie. (a) Farmer-reported scrapie (non-confirmed) from the 2002 anonymous postal survey, from which estimates for flock size were also obtained. (b) Confirmed cases of scrapie in SND in 2003, the year of the most recent breed survey [Reference Pollott and Stone26], from which estimates for flock size were obtained. (c) Confirmed cases of scrapie in SND from 2001 to 2005, the time-period covered by the NSP. Here, flock size was obtained from national breed surveys [Reference Pollott25, Reference Pollott and Stone26]. The risk of scrapie (s b) within breed b was estimated to be
where M bg is the proportion of sheep within the NSP of genotype g and breed b multiplied by the RCAM estimate, R g, for genotype g in the GB national flock [Reference Baylis3]. In each case, NSP data were restricted to the same time period as the scrapie incidence data being used.
RESULTS
Risk of scrapie for different genotypes within certain breeds
The estimated number of reported cases of scrapie per annum per million sheep (RCAM) and associated confidence intervals (CIs) were calculated for each PrP genotype in 17 breeds meeting the inclusion criteria for the study (Table 1). There was considerable variation in risk between PrP genotypes. The highest RCAMs within a breed tended to be in the VRQ/VRQ genotype. This was followed by the RCAMs for ARQ/VRQ (and ARH/VRQ in the case of the Texel); at 5–42% that of VRQ/VRQ, and the ARR/VRQ (0·2–12%) and ARQ/ARQ (0·3–10%) genotypes. These patterns agree with those obtained at the level of the national flock in which breed was not considered [Reference Baylis3].
Table 1. Estimates and 95% confidence intervals for number of reported cases of scrapie per annum per million sheep (RCAM) in each PrP genotype for 17 sheep breeds in GB

* Breeds are: Beulah Speckled Face (BSF); Bleu du Maine (BdM); Brecknock Hill Cheviot (BHC); Charollais (Cha); Cheviot (C); Easycare (E); Friesland (F); Greyface Dartmoor (GD); Herdwick (H); North Country Cheviot (NCC); Romney (R); Shetland (Sh); Suffolk (Su); Swaledale (Swa); Texel (T); Vendeen (V); Welsh Mountain (WM). Note: Suffolk sheep of the ARQ/ARQ genotype, previously considered the highest risk PrP genotype within this breed [Reference O'Rourke, Melco and Mickelson19, Reference Hunter30], have a risk of disease which is indistinguishable from other breeds.
White cells indicate no reported cases in a PrP genotype, despite the presumed presence of that genotype in a breed. Shaded cells indicate PrP genotypes believed to be absent or very rare in a breed [Reference Dawson2, Reference Eglin12, Reference Townsend, Warner and Dawson31]; and RCAMs shown in bold differ significantly (P<0·05) from the RCAM for the Texel sheep of the same PrP genotype.
Within a specific PrP genotype, there was also considerable variation in risk between certain breeds. For example, RCAMs for VRQ/VRQ animals ranged from 304 (North Country Cheviots), to 18 313 (Frieslands). RCAMs for ARQ/VRQ animals in the same two breeds ranged from 11 to 7631. These patterns suggest that some breeds have a higher incidence of scrapie (and hence a higher per capita risk) than others. It is noteworthy that Friesland sheep recorded the highest RCAMs of any breed in all four affected genotypes, while North Country Cheviots recorded the lowest in the two affected genotypes.
Of particular interest is the interaction between breed and genotype. For example, in the Shetland and Welsh Mountain breeds, the RCAMs for ARR/VRQ were higher than for ARQ/ARQ; the converse was true for Charollais, Cheviot, Swaledale and Texel. The RCAM in ARQ/ARQ sheep is about one three-hundredth and one-thirtieth that of VRQ/VRQ for the Shetland and Texel breeds, respectively. Taking the Texel breed as a baseline, some of these differences were statistically significant (Table 1). For example, ARQ/ARQ and ARQ/VRQ sheep of the Shetland breed had, respectively, significantly lower and higher RCAMs than their Texel counterparts.
Another result of interest is that in some breeds which lack the VRQ allele, scrapie is only reported in sheep in the ARQ/ARQ genotype (for instance, Suffolk and Vendeen), and while ARQ/ARQ is considered to be associated with a high risk of scrapie in these breeds [Reference Dawson2], the RCAM estimates suggest that the levels of risk are, in fact, similar to those of breeds that encode VRQ and experience only a small proportion of scrapie cases in ARQ/ARQ animals.
Flock-level variation in the PrP genotypes of cases
Distribution of PrP genotypes in affected flocks comprising multiple breeds
There were 12 flocks in which cases were confirmed in at least three breeds. Within most of these (Fig. 1a–d, f, g, k, l), the PrP genotypes of cases were largely consistent for all breeds within a single flock. A notable exception was flocks which included Texel sheep, where cases of disease were reported in ARH/VRQ animals; this is not surprising, as sheep of this genotype are rarely or never found in other breeds. While most or all breeds within a single flock tended to experience scrapie in similar PrP genotypes, there was evidence for differences in flocks. For example, the flock depicted in Fig. 1b experienced most scrapie in sheep lacking the VRQ allele; in the flocks depicted in Fig. 1(a, c, d, g–j, l), most cases encoded the VRQ allele; and in the flocks depicted in Fig. 1(e, f, k), there was a mixture of both.
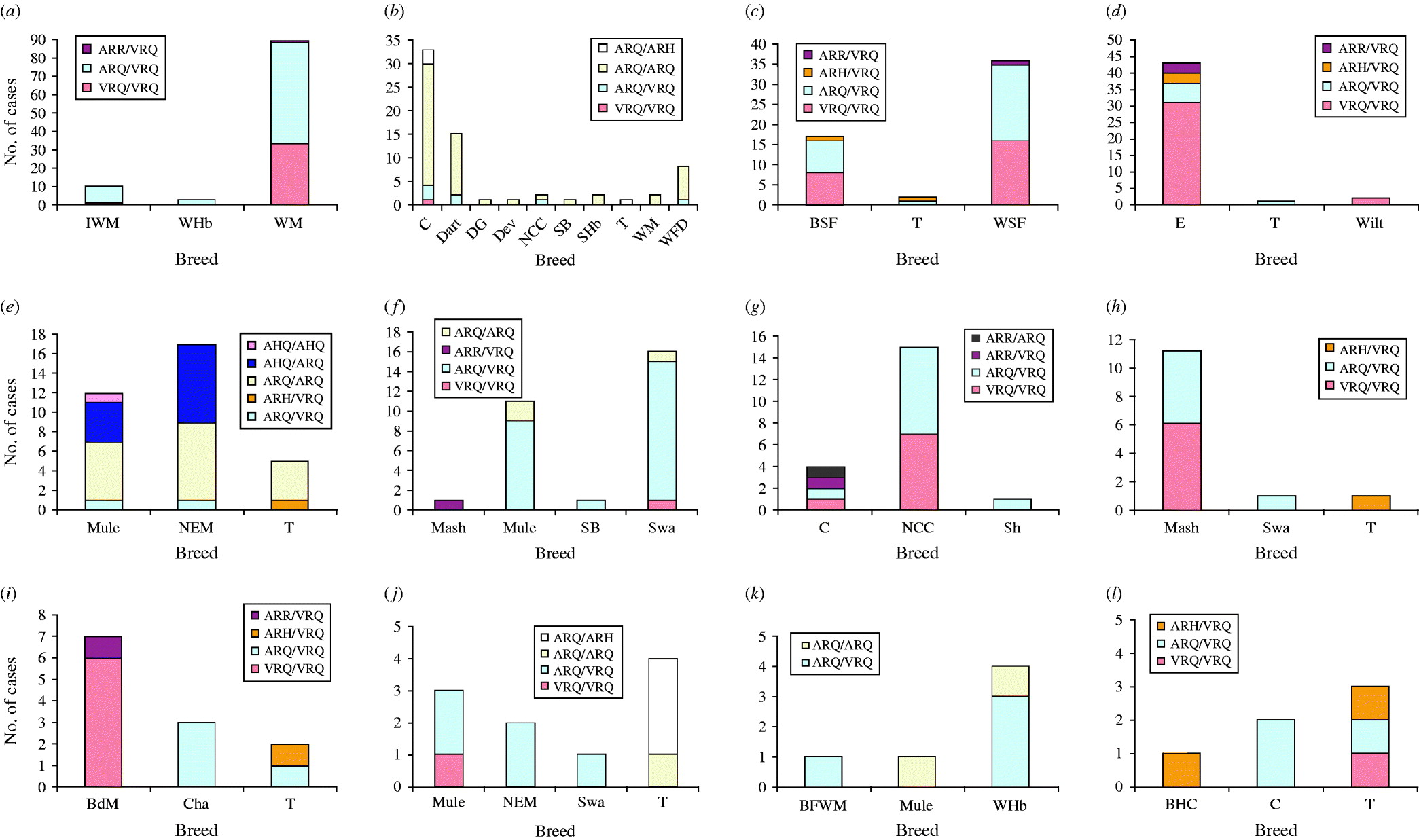
Fig. 1. Distribution of the PrP genotypes of confirmed cases in 12 single flocks (a–l) that comprised three or more distinct breeds of sheep. Breeds are: Badger Faced Welsh Mountain (BFWM); Beulah Speckled Face (BSF); Bleu du Maine (BdM); Brecknock Hill Cheviot (BHC); Charollais (Cha); Cheviot (C); Dartmoor (Dart); Derbyshire Gritstone (DG); Devon (Dev); Easycare (E); Improved Welsh Mountain (IWM); Masham (Mash); Mule (Mule); North Country Cheviot (NCC); North of England Mule (NEM); Scottish Blackface (SB); Scottish Halfbred (SHb); Shetland (Sh); Swaledale (Swa); Texel (T); Welsh Halfbred (WHb); Welsh Mountain (WM); Welsh Speckled Face (WSF); White Faced Dartmoor (WFD); Wiltshire (Wilt).
Distribution of PrP genotypes in breeds with multiple affected flocks
Six breeds were identified in which five or more cases were confirmed in two or more individual flocks (Friesland, Poll Dorset, Shetland, Swaledale, Texel, Welsh Mountain; Fig. 2). There were clear differences between breeds in the PrP genotypes of confirmed scrapie cases, which we attribute in part to the range of PrP genotypes encoded by each breed; for example, cases in ARH/VRQ were largely limited to Texels (Fig. 2e). With the exception of this genotype in Texels, the majority of cases were confirmed in animals of the VRQ/VRQ or ARQ/VRQ genotypes.

Fig. 2. Distribution of the PrP genotypes of confirmed cases in sheep breeds with at least five cases in two or more flocks. Breeds are (a) Friesland; (b) Poll Dorset; (c) Shetland; (d) Swaledale; (e) Texel; (f) Welsh Mountain.
While most flocks of a single breed experienced scrapie in the same genotypes, there was a small number of exceptions. For example, in one flock of Friesland sheep (Fig. 2a), two of six cases were in non-VRQ genotypes (AHQ/AHQ, ARQ/ARQ), while no cases were confirmed in these genotypes in three other Friesland flocks. In Swaledale sheep (Fig. 2d), four flocks had a high proportion of cases confirmed in ARQ/ARQ animals, while nine flocks had very few or none; this is despite the high frequency of the ARQ allele in the Swaledale breed, and the presumption, therefore, that ARQ/ARQ sheep were present in most or all of these flocks. Similar diversity is apparent in the eight affected Texel flocks (Fig. 2e). The array of affected genotypes indicates the presence of the ARQ, ARH and VRQ alleles in all eight flocks, but cases of disease in specific genotypes were only reported in a single (ARR/VRQ, ARH/ARH, VRQ/VRQ), two (ARQ/ARH) or three (ARQ/ARQ) flocks. By contrast, there was little or no evidence for between-flock variation in the genotypes of affected cases of the Poll Dorset, Swaledale and Welsh Mountain breeds (Fig. 2b, c, f).
The importance of PrP genotype frequency in the incidence of scrapie within breeds
Significant variation in the incidence of scrapie between 2001 and 2005 was explained by the estimated risk of disease within breeds (P=0·004), with a greater incidence when the risk within breeds was higher [adjusted odds ratio (aOR) 1·06, 95% CI 1·03–1·09]. This relationship was significant at the 10% level (P=0·06) for the incidence of scrapie in 2003 (aOR 1·05, 95% CI 1·00–1·10); however, breed risk did not explain variation in (unconfirmed) scrapie incidence indicated by the 2002 postal survey (P=0·3). Flock size did not explain variation in the incidence of scrapie (2001–2005, P=0·076; 2003, P=0·80; 2002, P=0·56) in any of the models.
DISCUSSION
By utilizing the largest data sources available for GB in this study, we have begun to tease apart the effects of PrP genotype and potential intrinsic differences between breeds in the risk of classical scrapie. Comparison of the estimated number of reported cases per annum per million sheep (RCAM) in PrP genotypes within different sheep breeds (Table 1) suggests, first, that some breeds have had more reported scrapie and, second, PrP genotype (as determined by the amino acids encoded by codons 136, 154 and 171) largely determines the risk of disease. In general, the ranking of PrP genotypes within each breed matched those for reported scrapie in the GB national flock as a whole [Reference Baylis3, Reference Tongue28].
Differences in the level of reporting amongst breeds and, hence, the risk of scrapie have been examined previously for cases reported between 1993 and 2002 (i.e. before the time-period considered in the present study), although the analysis did not include the effect of PrP genotype [Reference del Rio Vilas8]. Broadly similar patterns of relative risk were seen in that study as the present one, except for breeds predominantly farmed in Wales (Beulah Speckled Face, Brecknock Hill Cheviot, Welsh Mountain), all of which had a higher risk of scrapie than previously reported. Similarly, observations from anonymous postal surveys show that keeping certain breeds of sheep is a risk factor for scrapie, even after adjusting for other potential risk factors such as geographical location and husbandry practices [Reference McIntyre9, Reference McLean10]. Here we show for the first time that the frequency of PrP genotypes within breeds, weighted according to a national-level measure of risk, partly explains the incidence of confirmed clinical disease, such that those breeds with a higher frequency of high-risk PrP genotypes had a higher incidence of disease. Thus, differences in PrP genotype frequencies amongst breeds [Reference Eglin12] contribute to explaining observed differences in incidence.
Although the PrP genotype of an individual sheep appears to largely determine its risk of scrapie, there was nonetheless some evidence for differences in genotype-specific scrapie risk between certain breeds. This influence could reflect intrinsic differences in risk amongst breeds. However, it could also be a consequence of breed being confounded with other, extrinsic factors, most importantly scrapie strain. Sheep breeds tend to be associated with particular geographic locations, and flocks of a particular breed are more likely to interact with flocks of the same breed, with a corresponding increased risk of transmission; these factors will facilitate the circulation of distinct scrapie strains within specific sheep breeds and, so, scrapie strains that differentially target PrP genotypes and which circulate within specific breeds could lead to apparent breed-level differences.
Results presented here are consistent with a stronger influence of scrapie strain than sheep breed. While the PrP genotypes of cases are generally consistent within and amongst breeds (Table 1, Figs 1, 2), certain flocks provide notable exceptions where the PrP genotypes of cases are markedly different from those in other flocks of the same breed. These exceptions suggest that the strain of the scrapie agent may play an important role in determining affected PrP genotypes, but that most flocks of a single breed have the same or similar strains so that, in the absence of strain information, there is an apparent difference between breeds.
Scrapie strains circulating in the UK are poorly characterized, although limited strain typing in mice does suggest there is strain variation amongst flocks [Reference Bruce29]. The frequency of cases in VRQ-carrying animals suggests that most scrapie strains found in GB target the VRQ allele. Consequently a national-level measure (i.e. one which does not take sheep breed into account) is able to account for much of the risk of scrapie. Interestingly, the risk of disease in the ARQ/ARQ genotype is remarkably consistent across breeds, including breeds which lack the VRQ allele (Table 1). This suggests that the strains affecting these breeds could be, in effect, VRQ-attacking strains that are ‘trapped’ in breeds which lack this allele, as opposed to strains that are specifically adapted to targeting ARQ/ARQ or non-VRQ genotypes.
CONCLUSIONS
These investigations indicate that differences amongst breeds in the risk of classical scrapie play a less important role than either differences in the relative frequencies of PrP genotypes amongst breeds, or the strain of scrapie agent. Nevertheless, while targeting the VRQ allele in the NSP has prevented or limited the size of scrapie epidemics, there may be a small number of flocks that harbour strains which attack other alleles, and which could seed other outbreaks if not resolved. The design of the NSP which focuses on PrP genotype alone is reasonable, although in certain circumstances it may become marginally more efficient if it considered breed, and with an even stronger focus, scrapie strain.
ACKNOWLEDGEMENTS
This work was funded by the Biotechnology and Biological Sciences Research Council (grant no. IAH1320). H.T. was supported by a summer student bursary from the IAH. The authors are grateful to Mike Dawson (NSP Advisory Centre) for supplying PrP genotype data and Victor del Rio Vilas (Veterinary Laboratories Agency) for supplying data on reported cases.
DECLARATION OF INTEREST
None.





