INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) infections are an important cause of morbidity and a major public health problem in the USA [Reference Klevens1]. Although MRSA was traditionally considered a healthcare-associated pathogen, MRSA has emerged worldwide as an important cause of community-associated skin-and-soft-tissue infections [Reference Vandenesch2]. In the USA, MRSA pulsed-field gel electrophoresis (PFGE) type USA300 strains have been responsible for the majority of community-associated MRSA infections [Reference Moran3]. Clusters of community-associated MRSA infections have been described in incarcerated adults [4–Reference Turabelidze7], persons participating in team sports [Reference Lindenmayer8–Reference Kazakova11], military recruits [Reference Zinderman12], and sexually active men who have sex with men (MSM) [6, Reference Lee13].
High rates of both community-associated [Reference Skiest14–Reference Crum-Cianflone, Burgi and Hale16] and healthcare-associated MRSA infections have also been described in HIV-infected individuals, although the underlying basis for this association is unknown. Proposed mechanisms include immune dysfunction [Reference Mathews15, Reference Thompson and Torriani17], behavioural risk factors [Reference Lee13], and increased exposure to the healthcare system [Reference Bozzette18]. Colonization with S. aureus is a risk factor for subsequent clinical infection [Reference Nguyen19, Reference von Eiff20] and the site of colonization may also be an important risk factor for clinical infection [Reference Szumowski21]. Although the anterior nares is considered the primary reservoir of S. aureus [Reference Wertheim22], MRSA PFGE type USA300 might preferentially colonize the buttocks, genitals and perineum [Reference Szumowski21] leading to more infections in these anatomical areas due to this type. The prevalence of MRSA colonization in HIV-infected individuals is high (10–17%) [Reference Cenizal23, Reference Hidron24] compared to the general USA population (0·8–1·5%) [Reference Gorwitz25, Reference Kuehnert26]. Improving our understanding of the interaction between HIV infection and MRSA colonization is necessary in order to develop effective strategies to prevent MRSA infections in this population. We, therefore, conducted this study to determine both the prevalence of and risk factors for MRSA colonization in HIV-infected adults.
METHODS
Study design
Study participants were recruited from the Atlanta Veterans Affairs Medical Center (VAMC) HIV clinic, which provides medical care to about 1200 HIV-infected veterans and is the largest VAMC HIV clinic in the USA. This study was approved by the Emory University and Centers for Disease Control and Prevention (CDC) Institutional Review Boards and the VA Research and Development Committee. Eligible participants were HIV-infected, aged ⩾18 years, receiving outpatient medical care at the Atlanta VAMC HIV clinic, and competent to provide informed consent. All eligible participants who attended the clinic from September 2007 to April 2008 were invited to participate in the study by a study coordinator or were given a flyer with relevant study details and contact information. All participants provided informed consent to take part in this study and completed the study visit on the same day as their clinic visit.
Data on demographic characteristics, medical history, antibiotic use within the past 12 months, and microbiological data on previous S. aureus infections were obtained from electronic medical records. The most recent CD4 cell count and viral-load results were used at study enrolment. Participants also completed a questionnaire that focused on their living situation (residential, nursing home, correctional facility, homeless), self-reported history of skin infections, personal hygiene, sexual behaviour, and drug use over the past 12 months. All questionnaires were completed in private and all study data were anonymized to ensure confidentiality.
Microbiological procedures
Samples for S. aureus culture were collected from the anterior nares and the groin at study enrolment using a sterile rayon swab and placed in Liquid Stuart's transport media (Becton Dickinson, USA). Study staff sampled the anterior nares but participants were instructed to self-sample the groin by swabbing in the skin folds between the thigh and genital area. Swabs were plated on mannitol salt agar (Becton Dickinson) and CHROMagar™ MRSA (Becton Dickinson) and then placed in 5 ml trypticase soy broth with 6·5% sodium chloride (Becton Dickinson). Mannitol salt agar plates were examined after 48-h incubation at 35°C, and mannitol-fermenting colonies with a distinct morphotype were subcultured on a trypticase soy agar plate with 5% sheep blood (Becton Dickinson). Inoculated trypticase soy broth was incubated for 24 h at 35°C and then subcultured to mannitol salt agar and processed as above. CHROMagar plates were incubated at 35°C for 48 h and were examined after 24 h and 48 h for the presence of mauve-coloured colonies. S. aureus was identified by morphology and either a positive Staphaurex® latex agglutination test (Remel, USA) or a positive tube coagulase test. Methicillin susceptibility was determined with a cefoxitin disk-diffusion test according to Clinical and Laboratory Standards Institute (CLSI) guidelines [27].
All MRSA isolates were typed by PFGE with SmaI (New England Biolabs, USA) as described previously [Reference Gorwitz25], using Salmonella enterica serovar Braenderup H9182 as the normalization standard. PFGE patterns were analysed with BioNumerics software version 5.10 (Applied Maths, USA) and isolates were grouped into types using Dice coefficients and 80% relatedness [Reference McDougal28]. MRSA PFGE types USA500 and Iberian were grouped together (USA500/Iberian) for this analysis because they are closely related and difficult to separate by PFGE [Reference McDougal28]. Antimicrobial susceptibility testing of all MRSA isolates for resistance to oxacillin, cefoxitin, penicillin, chloramphenicol, clindamycin (both constitutive and inducible), erythromycin, daptomycin, doxycycline, tetracycline, gentamicin, levofloxacin, linezolid, mupirocin, rifampin, trimethoprim–sulfamethoxazole (TMP–SMX) and vancomycin was performed by reference broth microdilution and interpreted according to CLSI guidelines [27]. PCR was used to screen for staphylococcal cassette chromosome mec types (SCCmec) II and IV and to detect Panton–Valentine leukocidin (PVL) genes in MRSA PFGE type USA300 isolates [Reference Limbago29].
Statistical methods
The primary outcome in this analysis was MRSA colonization. Participants were classified as MRSA-colonized if MRSA was detected from the mannitol salt agar or trypticase soy broth cultures of the nares or groin. Participants were classified as colonized with methicillin-susceptible S. aureus (MSSA) if MSSA was detected and MRSA was not detected. Participants colonized with both MSSA and MRSA (regardless of site of colonization) were classified as MRSA-colonized. Participants were classified as not colonized with S. aureus if neither MRSA nor MSSA were detected. The primary analysis compared participants with MRSA colonization with those not colonized with MRSA (MSSA or no S. aureus). A secondary analysis compared participants carrying MSSA with those negative for S. aureus. Of MRSA-colonized participants we also compared USA300 colonization with colonization from other MRSA PFGE types. All analyses were performed using SAS version 9.1 (SAS Institute Inc., USA). The Wilcoxon rank-sum test (continuous variables) and the χ2 and Fisher's exact tests (categorical variables) were used to test for differences in clinical, demographic, and behavioural variables (e.g. CD4 cell count, living situation) in participants with and without MRSA. Statistically significance was indicated by a P value <0·05. Using multivariate logistic regression (SAS Proc Logistic), adjusted odds ratios (aOR) and 95% confidence intervals (CI) were calculated to identify variables associated independently with outcomes. All statistically significant variables in univariate analysis (exact unadjusted OR) were included in a multivariate model [Reference Altman30] and variables with P>0·05 were dropped sequentially to create a parsimonious model that was manually reviewed after each step. An F test was also performed using the Type III sum of squares of the dropped variables from the saturated model to verify that the latter were not important to the model. The model was also evaluated with the Hosmer & Lemeshow goodness-of-fit test.
RESULTS
Overall 600 (95%) of 629 HIV-infected veterans who were provided information by a study coordinator elected to participate. Nares and groin samples were obtained from these 600 participants (~50% of the clinic population), the majority were male (98%) with a median age of 52 years [interquartile range (IQR) 45–59 years]. Four hundred and forty-one (74%) participants were non-Hispanic black and 315 (53%) reported being a man who had sex with men (MSM) as their HIV transmission risk factor. The median most recent CD4 cell count was 416 cells/μl (IQR 250–579 cells/μl) and 474 (79%) participants were receiving antiretroviral therapy (ART). Participants were not significantly different from HIV-infected veterans who did not take part in this study with respect to gender, age, HIV transmission risk factor, and proportion receiving antiretroviral therapy (data not shown).
MRSA colonization was detected in 79 (13%) participants, including three who were colonized with two distinct MRSA strains (82 total MRSA isolates), and MSSA colonization was detected in 180 (30%) participants (Fig. 1). USA300 (44/82, 54%) and USA500/Iberian (29/82, 35%) were the most common MRSA PFGE types identified. MRSA was detected solely in the nares for 30 (38%) individuals, in both the nares and groin for 36 (46%), and only in the groin for 13 (16%) participants (Fig. 2). USA300 was identified in 11/30 (37%) participants with MRSA solely in the nares, 21/36 (58%) with MRSA in both the nares and groin (including the three co-colonized), and 11/13 (85%) with MRSA solely in the groin. USA300 was more likely to be detected only in the groin (P=0·04) compared to USA500/Iberian. Including groin cultures increased the overall detection of MRSA by 24% and increased the yield of USA300 detection by 38% compared to nasal culture alone. Of those colonized with MSSA only (n=180), MSSA was detected in the nares alone for 74 (41%) participants, in both the nares and groin for 83 (46%), and in the groin alone for 23 (13%) participants.
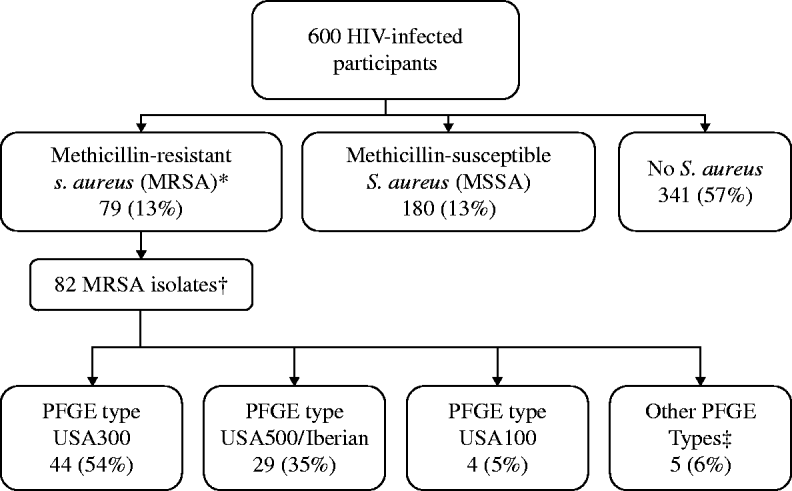
Fig. 1. Prevalence of Staphylococcus aureus recovered from nares and groin swabs in HIV-infected adults. * Eleven participants were co-colonized with MRSA and MSSA. These participants were classified as MRSA-colonized. † Three participants were co-colonized with MRSA pulsed-field del electrophoresis (PFGE) type USA300 and a second PFGE type: USA100, USA500, USA1000 (n=1 each). ‡ USA700 (n=1), USA800 (n=2), and USA1000 (n=2).
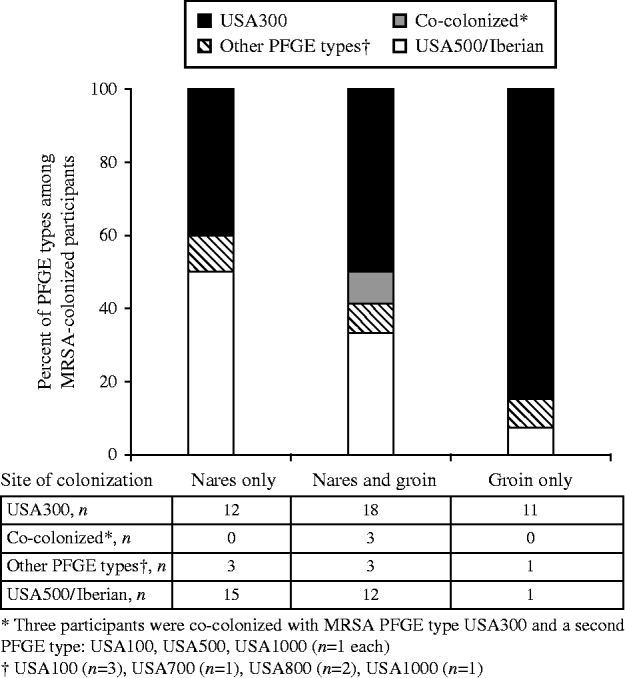
Fig. 2. Proportion of pulsed-field gel electrophoresis (PFGE) types by anatomical site of detection in MRSA-colonized HIV-infected adults (n=79).
In univariate analysis, study subjects with MRSA colonization were similar to those with no MRSA colonization with respect to age, race/ethnicity, HIV transmission risk factors, and drug use in the past 12 months (Table 1). MRSA colonization was associated with a lower CD4 cell count (<500 cells/ml), a previous history of an abscess, a past medical history of MRSA clinical infection, hospitalization within the past 12 months, a history of syphilis infection, the use of certain anti-staphylococcal agents in the past 12 months, residing or working in a prison or jail, and reporting rarely or never using condoms during sex. MRSA colonization was detected in 18/27 (67%) participants with a clinical MRSA infection within the past year, 8/18 (44%) with a clinical MRSA infection from 1–2 years prior to enrolment, and 10/36 (28%) participants with a clinical MRSA infection >2 years prior to enrolment. Although MRSA colonization rates declined with time since the clinical MRSA infection, these rates remained high compared to participants who never had an MRSA infection [MRSA colonization was detected in 43 (8%) of 519 participants without a history of an MRSA clinical infection].
Table 1. Factors associated with MRSA colonization in HIV-infected adults (n=600)
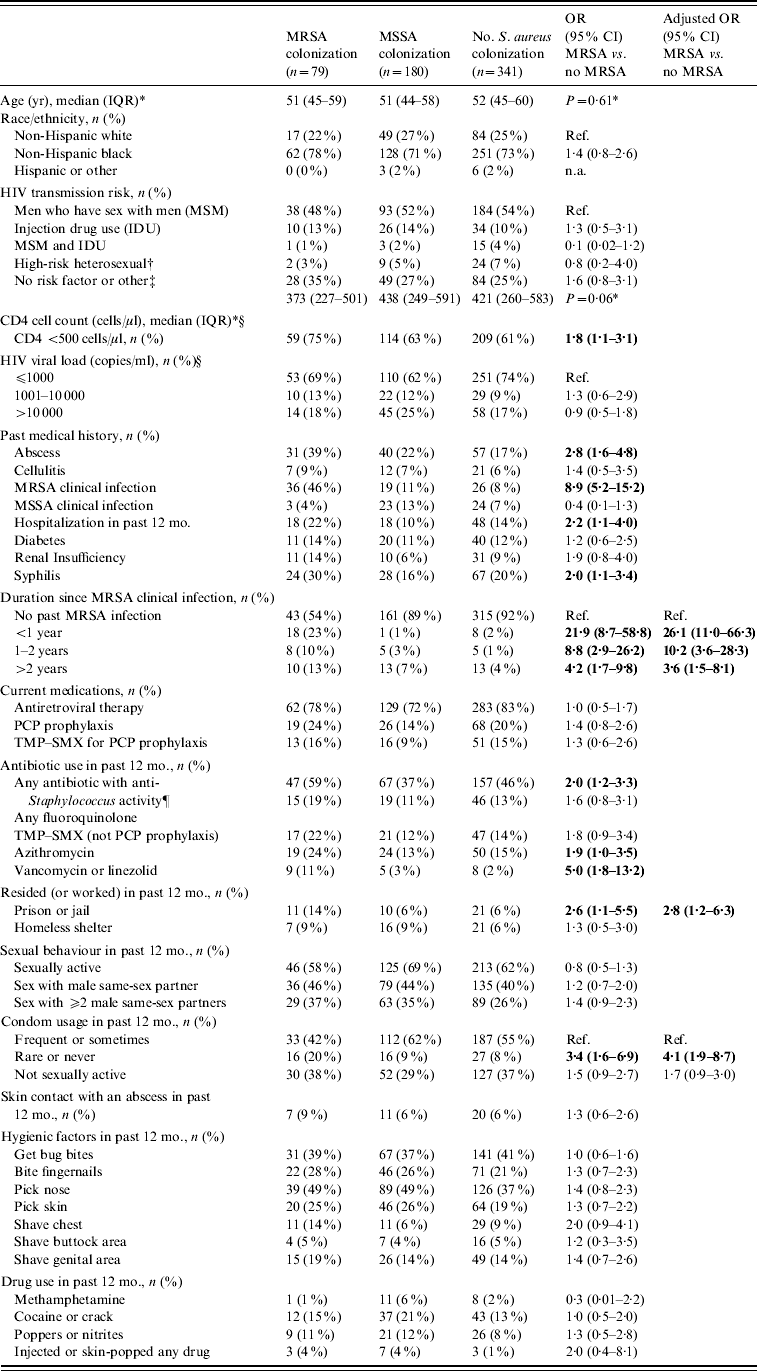
OR, Odds ratio; CI, confidence interval; TMP–SMX, trimethoprim–sulfamethoxazole; PCP prophylaxis, Pneumocystis pneumonia prophylaxis.
* Values are medians with interquartile ranges.
† Sexual contact with a person known to be HIV-infected or at high risk for HIV infection (e.g. history of IDU or MSM).
‡ Risk not specified or unknown (n=152), ‘Other’ (n=9) included Transfusion (n=4), Healthcare worker occupational exposure (n=5).
§ CD4 cell count and viral load results were the most recent CD4 cell count and viral load results at the time of the nares and groin cultures.
¶ Antibiotics with anti-staphylococcal activity used were fluoroquinolones, TMP–SMX, azithromycin, clarithromycin, mupirocin, cephalexin, vancomycin, linezolid, and ampicillin–clavulanate.
In multivariate analysis, participants colonized with MRSA were significantly more likely to have had a past medical history of MRSA clinical infection, to have reported rarely or never using condoms, and to have resided or worked in a prison or jail (Table 1). In a multivariate model that excluded 81 participants with a past medical history of MRSA clinical infection, residence or work in a prison or jail (aOR 2·9, 95% CI 1·0–7·7), rarely or never using condoms (aOR 4·3, 95% CI 1·7–10·4), and a history of syphilis infection (aOR 2·4, 95% CI 1·1–5·0) remained independently associated with an increased risk of MRSA colonization.
The univariate analysis was repeated comparing separately MRSA-colonized with MSSA-colonized participants and those negative for S. aureus; the same associations listed above were observed (data not shown). In addition those colonized with MSSA were similar to S. aureus-negative subjects with respect to age, race/ethnicity, HIV transmission risk factors, CD4 cell count, and the use of certain anti-staphylococcal agents in the past 12 months. Compared to no S. aureus colonization, MSSA carriers were associated with drug use in the past 12 months, a higher viral load, a past medical history of MSSA clinical infection, not receiving antiretroviral therapy, nose picking, and having sex with ⩾2 partners in the past 12 months (Table 2).
Table 2. Univariate analysis of factors associated with MSSA colonization in HIV-infected adults (n=521Footnote *)
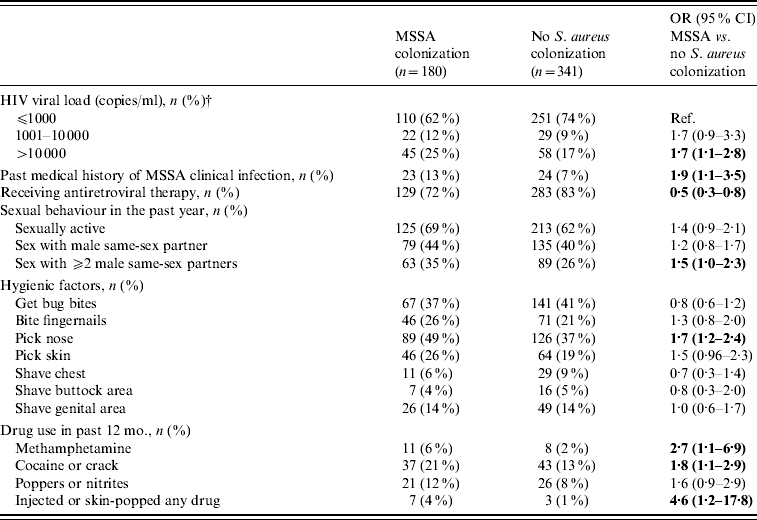
OR, Odds ratio; CI, confidence interval.
* This analysis excludes the 79 participants with MRSA colonization.
† Viral load results were the most recent viral load results at the time of the nares and groin cultures.
Of the 44 USA300 isolates 31 (70%) carried SCCmec IVa and PVL; the remaining 13 lacked PVL (12 were MRSA variant pattern USA300-0045 and six carried SCCmec IVb). Of USA300 isolates encoding PVL, resistance to levofloxacin (14/31, 45%), clindamycin (5/31, 16%), and tetracycline (7/31, 23%) was observed but at lower rates compared to other PFGE types (Table 3). Resistance to linezolid, doxycycline, or vancomycin was not detected.
Table 3. Antimicrobial resistance profiles of MRSA and PFGE types in HIV-infected adults (n=82 isolatesFootnote *)
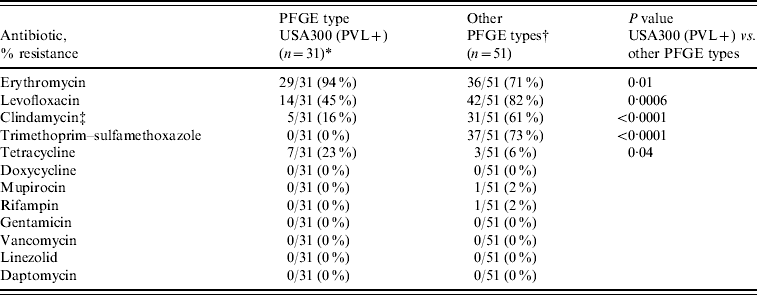
* MRSA PFGE type USA300 encoding PVL genes.
† Including USA500/Iberian (n=29), USA300 without PVL genes (n=13), USA100 (n=4), USA800 (n=2), USA1000 (n=2), and USA700 (n=1).
‡ MRSA colonization was detected in 79 (13%) participants, including three who were colonized with two distinct MRSA strains, resulting in 82 isolates.
In a multivariate analysis limited to participants with MRSA, colonization with USA300 encoding PVL was associated with a higher CD4 cell count (⩾500 cells/μl) and a self-reported history of a boil or abscess (Table 4). However, participants colonized with USA300 encoding PVL were similar to subjects with other MRSA PFGE types with respect to a past medical history of MRSA clinical infection, residence or work in prison or jail, rarely or never using condoms, and a history of syphilis infection. To explore these associations further, the overall prevalence of colonization with USA300 encoding PVL and other MRSA PFGE types were stratified by CD4 cell count and a self-reported history of a boil or abscess. Participants with a CD4 cell count ⩾500 cells/μl had a similar prevalence of colonization with USA300 encoding PVL (6% vs. 4·5%, P=0·66) but a lower prevalence of other MRSA PFGE types (3% vs. 10·5%, P=0·001) compared to a CD4 cell count <500 cells/μl. Participants with a self-reported history of a boil or abscess had a higher prevalence of colonization with USA300 encoding PVL (12% vs. 3%, P<0·0001) and a similar prevalence of other MRSA PFGE types (8% vs. 8%, P=0·70) compared to participants without a self-reported history of a boil or abscess.
Table 4. Factors associated with MRSA colonization with PFGE type USA300 encoding PVL genes in HIV-infected adults (n=79)
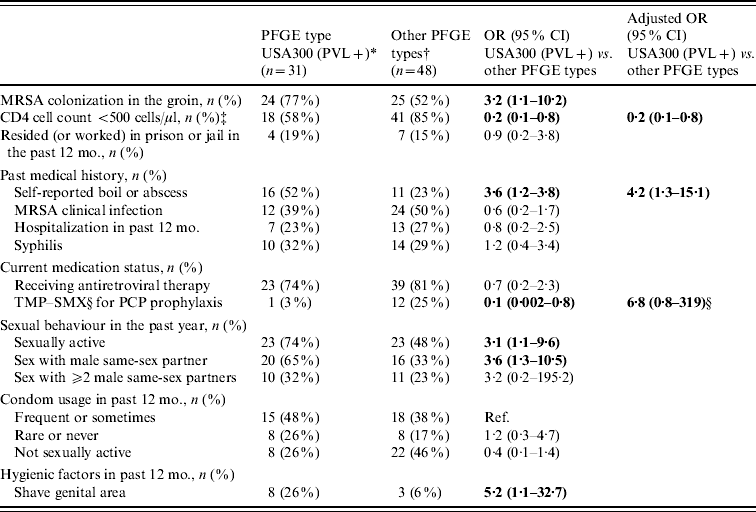
TMP–SMX, Trimethoprim–sulfamethoxazole; PCP prophylaxis, Pneumocystis pneumonia prophylaxis.
* MRSA PFGE type USA300 encoding PVL genes.
† Including USA500/Iberian (n=28), USA300 without PVL genes (n=13), USA100 (n=3), USA800 (n=2), USA1000 (n=1), and USA700 (n=1).
‡ CD4 cell count results were the most recent CD4 cell count results at the time of the nares and groin cultures.
§ TMP–SMX for PCP prophylaxis was left in the multivariate model because its use is related to CD4 cell count.
DISCUSSION
The prevalence of MRSA colonization was high (13%) in this study of HIV-infected adults compared to the general population [Reference Gorwitz25, Reference Kuehnert26]. USA300 and USA500/Iberian accounted for the majority of MRSA-colonizing isolates. Inclusion of groin cultures increased the yield of USA300 detection by 38% compared to culturing of the nares alone. A past medical history of a clinical infection with MRSA conferred the greatest risk for MRSA colonization. Correlates of risky sexual behaviour (i.e. rarely or never using condoms) and contact with jails and prisons were also independently associated with MRSA colonization.
In the general USA population, the prevalence of MSSA colonization ranges from 27·2% to 30·0% [Reference Gorwitz25, Reference Kuehnert26] and MRSA colonization is uncommon (0·8–1·5%) although there is evidence that it is increasing [Reference Gorwitz25, Reference Kuehnert26]. In this study of HIV-infected adults, the prevalence of MSSA colonization was similar to the general population prevalence (30%) but the prevalence of MRSA colonization was markedly higher (13%). The high prevalence of MRSA colonization we observed is consistent with previous studies of HIV-infected hospitalized patients in Atlanta (17%) [Reference Hidron24] and HIV-infected outpatients in New York City (17%) [Reference Shet31] and in Texas (10%) [Reference Cenizal23] but higher than rates reported in HIV-infected outpatients from Boston (4%) [Reference Szumowski21], Nebraska (2%) [Reference Madariaga, Ullrich and Swindells32], Brazil (0%) [Reference Padoveze33], Germany (2%) [Reference Seybold34], and Italy (0%) [Reference Giuliani35]. This wide variation in prevalence suggests that HIV-infected adults are highly susceptible to MRSA colonization but that the absolute prevalence may depend on local factors, such as the intensity of environmental exposure to MRSA.
Prior studies have demonstrated that USA300 causes most community-associated MRSA infections in the USA [Reference Klevens1, Reference Moran36], whereas USA500/Iberian clones are associated with healthcare-associated MRSA infections [Reference Klevens1]. The equal distribution of PFGE types USA300 and USA500/Iberian in our study suggests that community-associated and healthcare-associated exposures were both important determinants of MRSA colonization in this population. Interestingly, evolutionary studies suggest that USA300 and USA500 evolved from a common ancestor [Reference Li37] suggesting that these strain types may share genetic determinants that favour colonization in this population. USA100, traditionally the most common healthcare-associated MRSA PFGE type in the USA [Reference Klevens1], was isolated infrequently in our study and its relative absence might reflect the predominant type of healthcare exposures (outpatient clinic visits) that our participants experienced.
MRSA colonization was more frequent in participants with a CD4 cell count <500 cells/μl (in univariate analysis) and these participants had a higher prevalence of colonization with MRSA PFGE types other than USA300 encoding PVL (including healthcare-associated PFGE types such as USA500/Iberian). Although MRSA infection is not a traditional HIV-associated opportunistic infection, immunological dysfunction related to HIV infection, such as a low CD4 cell count [Reference Ramsetty38] or impaired neutrophil function [Reference Pugliese39], could increase susceptibility to MRSA colonization. CD4 cell count may also be a marker for differences in exposures that were not measured in our study. For example, individuals with low CD4 cell counts may have more interactions with the healthcare system and more exposures to healthcare-associated MRSA PFGE types (USA500/Iberian). We also observed that USA300 colonization (irrespective of PVL status) was more frequently detected in the groin than in the nares. Although the anterior nares is considered the primary reservoir of S. aureus [Reference Wertheim22], USA300 might preferentially colonize the buttocks, genitals and perineum [Reference Szumowski21] leading to more USA300 MRSA infections in these anatomical areas. Understanding the mechanisms that allow USA300 to colonize and cause disease at these sites will be an important step in developing more effective strategies to prevent these infections.
Participants receiving TMP–SMX prophylaxis were less likely to have colonization with USA300 encoding PVL (in univariate analysis), consistent with the observation that this PFGE type is usually susceptible to TMP–SMX [Reference Moran36]. Although we observed less resistance to clindamycin and levofloxacin in USA300 isolates compared to USA500/Iberian, the presence of any resistance to these therapeutic options is of concern. A study in MSM in San Francisco and Boston documented a high prevalence of community-associated infections caused by USA300 isolates encoding SCCmec IVa and PVL that contained a conjugative plasmid that carried resistance elements for macrolides, clindamycin, and mupirocin [Reference Diep40]. In our study, we did detect resistance to TMP–SMX in USA300 isolates that lacked PVL genes and carried SCCmec IVb (MRSA PFGE pattern USA300-0045). However, these isolates are genetically distinct from the USA300 isolates that have caused the majority of community-associated MRSA infections in the USA and their relative contribution to MRSA clinical infections in HIV-infected individuals is unknown. Continued surveillance for antimicrobial resistance in USA300 isolates is necessary to determine if antimicrobial resistance is increasing.
A past medical history of microbiologically confirmed MRSA clinical infection was a major risk factor for MRSA colonization; almost half of the MRSA-colonized participants in our study had a history of MRSA clinical infection. With cross-sectional data it is unclear if MRSA colonization preceded clinical infection or vice versa. However, the decline in prevalence of MRSA colonization with increasing time since clinical infection suggests that clinical infection might precede colonization in this population. Alternatively MRSA colonization could precede clinical infection and persist despite resolution of the clinical infection. We also found that certain behavioural and environmental risk factors, such as rarely or never using condoms and contact with jails and prisons, were associated with risk for MRSA colonization but not risk for MSSA colonization. This suggests that MRSA has established its own ecological niche and is not simply replacing MSSA, a theory supported by other studies [Reference Hota41].
There are several limitations to our study. First, our study population was 98% male and our findings are not generalizable to HIV-infected women. Second, some risk factors for community-associated MRSA clinical infections, such as methamphetamine use [Reference Lee13] and close contact with someone with a skin infection [Reference Moran36], were not associated with MRSA colonization in our study. These differences might reflect different risks for MRSA infection and colonization or might be explained by low frequencies of certain risk factors (i.e. methamphetamine use) in our study. Third, we have interpreted rarely or never using condoms as representing risky sexual behaviour which, however, may simply be a marker of general hygiene or reflect an increased likelihood of having skin-to-skin contact with a partner who has an active MRSA infection. Other markers of risky sexual behaviour (e.g. sex with ⩾2 partners in the past year) were not associated with MRSA colonization in our study. Finally, our hypothesis that most USA300 was acquired in the community and most USA500/Iberian was acquired in a healthcare setting is also speculative and could be explored further with environmental sampling.
In conclusion, MRSA colonization was common in this study of HIV-infected adults. MRSA PFGE types USA300 and USA500/Iberian both contributed to colonization suggesting the importance of both community-associated and healthcare-associated exposures. Given this high burden of colonization, community-associated MRSA prevention strategies (such as keeping cuts and scrapes clean and covered, practising good hand hygiene, avoiding shared personal items such as towels and razors, and decolonization in certain situations [Reference Gorwitz42]) should be emphasized in HIV-infected adults in communities with high rates of MRSA clinical infections. Colonization with USA300 was frequently found only in the groin, and preventive strategies will need to consider the high frequency of USA300 colonization at this anatomical site.
ACKNOWLEDGEMENTS
This publication was made possible by support from the Division of HIV/AIDS Prevention and the Division of Healthcare Quality Promotion, CDC. The findings and conclusions in this report are those of the authors and do not necessarily represent the official views of the CDC.
DECLARATION OF INTEREST
None.







