INTRODUCTION
Verocytotoxin-producing Escherichia coli (VTEC) are regarded as emerging pathogens that cause several gastrointestinal illnesses in humans and animals, ranging from mild diarrhoea to much more severe diseases such as haemorrhagic colitis (HC) or haemolytic uraemic syndrome (HUS) [Reference Sherwood, Snodgrass and O'Brien1–Reference Chart5]. Cases of human VTEC infection are increasingly common with serogroup O157 being the dominant type in the United Kingdom and Northern America [Reference Willshaw2, Reference Jones6, Reference Parry and Palmer7], while other serogroups such as O26, O103, O111 and O145 have also been isolated [Reference Willshaw8]. Most human VTEC infections are thought to arise from contacts with contaminated sources such as farm animals, pets, food and water [Reference Willshaw2, Reference Ryan9–Reference Trevena12]. This finding, together with the high VTEC prevalence within cattle herds [Reference Chapman13, Reference Mechie, Chapman and Siddons14] and the association between human cases and farm densities [Reference Kistemann15], suggest that cattle are potential reservoirs for human VTEC infections. However, the sources and the epidemiological dynamics of different VTEC within cattle populations are mostly unclear and therefore require further investigation.
Given the importance of VTEC in causing human and animal diseases, several methodologies have been developed for their recovery from contaminated sources including PCR–DNA probe hybridization [Reference Willshaw2, Reference Shaw16], immunomagnetic separation (IMS) [Reference Chapman and Siddons17, Reference Cubbon18] and monoclonal ELISA [Reference Randall, Wray and Mclaren19]. The sensitivity of VTEC detection, which can be defined as the probability of an infected sample testing positive, varies between different approaches from as low as 55% to a much higher level of 94% [Reference Capps20]. Thus, it is important to investigate whether the issue of test sensitivity affects our assessment of the epidemiological characteristics of VTEC.
Motivated by the above, longitudinal studies were carried out on two Scottish beef farms to monitor the spread of VTEC serogroups O26 and O103 in different calf cohorts. In order to investigate the sensitivity issue two methods were used for isolating serogroup O26 in one of the farm studies. A subset of VTEC-positive samples from one of the calf cohort studies also had their PFGE types determined. The objectives of this paper are thus: (1) to fit mathematical models to the observed data and then estimate epidemiological parameters relating to the duration and transmissibility of VTEC O26 and O103 infections; (2) to quantify the sources of individual infections; (3) to compare the epidemiological characteristics of VTEC O26 and O103 within the same calf cohort studies, and the same VTEC serogroups in two different calf cohorts; (4) to fit mathematical models to different datasets obtained from different detection methods for serogroup O26 in the same cohort, and see how our conclusions regarding the epidemiological characteristics of serogroup O26 can be affected by test sensitivity; and (5) to determine whether the heterogeneity in the PFGE types of isolates reflects that in the environment, or is generated via the calf-to-calf transmission processes.
METHODS
Data description
Two calf cohort studies, CC1 and CC2 (representing the first and second calf cohort studies respectively), were carried out on two Scottish beef farms in autumn 2001 and spring 2004 respectively. In CC1, faecal samples were taken from 49 calves once a week from birth for 17 weeks. A slide agglutination test with TBX agar was used to detect VTEC serogroups O26 (hereafter CC1 TBX O26) and O103 (CC1 TBX O103) in all faecal samples. Of a total of 570 faecal samples collected, 100 were positive for serogroup O26 and 27 were positive for serogroup O103 (Fig. 1).

Fig. 1. VTEC serogroups O26 and O103 isolated from calves plotted by sampling week during the course of both calf cohort studies. In the figure, CC1 and CC2 represent the first and the second calf cohort studies respectively; TBX and Rh are the agglutination tests with TBX agar and rhamnose agar respectively. Each box represents a sample taken from a particular calf (identified by the labels on the vertical axis) on a particular week (identified by the week number on the horizontal axis). Empty boxes represent samples with no VTEC serogroups detected. Black boxes represent samples with VTEC serogroups detected using the agglutination test with TBX agar. In the figure CC2 TBX O26, boxes representing samples testing positive for VTEC O26 using the rhamnose agar, are shaded in grey; and samples testing positive for VTEC O26 by using TBX and rhamnose agars are coloured in black with grey shaded background.
In CC2, faecal samples were taken from 41 calves once a week from birth for 19 weeks. A slide agglutination test with TBX agar was used to identify VTEC serogroups O26 (CC2 TBX O26) and O103 (CC2 TBX O103). Of a total of 686 samples collected, 31 were positive for serogroup O103 and 154 for serogroup O26 (Fig. 1). Another slide agglutination test with rhamnose agar, which is known to have good sensitivity and specificity for isolating VTEC serogroup O26 [Reference Hiramatsu21], was also used to detect serogroup O26 (CC2 Rh O26). It was found that 281 out of 686 samples collected were positive (Fig. 1). Furthermore, 134 out of 281 O26-positive samples isolated with rhamnose agar failed to be detected with TBX agar and seven out of 154 O26-positive samples isolated with TBX agar failed to be detected with rhamnose agar.
In CC2, all isolates of VTEC serogroups O103 had their PFGE patterns determined by using methods described previously (see [Reference Vali22]). For serogroup O26, PFGE patterns were also obtained for a sub-collection of samples testing positive for serogroup O26 using both TBX and rhamnose agars. For VTEC serogroup O103, three different PFGE patterns were characterized and there was a single dominant type (Table 1). For VTEC serogroup O26, 12 different PFGE patterns were characterized and there were two dominant types (Table 1).
Table 1. A summary of frequencies of different PFGE types found in VTEC-positive samples in the second calf cohort study
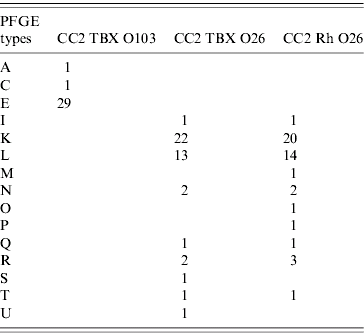
The labelling of PFGE types is for the ease of identification and has no biological meaning. The identities of individual datasets and their abbreviations are as explained in the main text.
Basic model and parameter estimation
We adopt the same methodology used previously [Reference Liu23] to construct mathematical models and estimate parameter values. We briefly describe the methodology here. First, we construct a stochastic susceptible–infected–susceptible (SIS) model for a VTEC serogroup with cohort structure. A susceptible calf (S) can acquire infection via two routes. The first is from another infected calf (I) (including direct and indirect contacts) at a rate βI, where β is the within-cohort transmission coefficient. The within-cohort infection term is considered to be a function of the absolute number of infectious calves in the limited area occupied by the cohort and we therefore have modelled the infection process in a density-dependent manner. Second, a susceptible calf can acquire infection from sources other than infected calves at a rate θ. Such environmental sources include other infected cattle, animals and other contaminated objects. An infected calf can recover at a rate γ to become susceptible. The basic outline of the SIS process is summarized in Figure 2 and can be described by the following set of differential equations:
Simulations of the model are used to generate samples from the population at intervals according to the actual sampling frame employed in CC1 and CC2. Maximum-likelihood methods are used to estimate the parameter values and associated confidence intervals [Reference Hilborn and Mangel24, Reference Hudson25]: this involves systematically changing parameter values; and with every parameter set 1 000 000 model simulations are generated; we then count how frequently the model reproduces the observed data. The probability of observing the data, given the model and parameter values, is then calculated. The best set of parameter values is the one with the highest probability of reproducing the data. Following our previous approach [Reference Liu23], we are required to define some properties of the data to allow comparison between model output and the observed data. These are: (1) the number of positive infections, which is the number of samples that tested positive for a particular VTEC serogroup using a particular detection method; (2) the infection week, which is the number of weeks during which at least one sample tested positive; and (3) the number of animals that ever tested positive for a particular serogroup. Table 2 summarizes the three properties for the data in both calf cohort studies.

Fig. 2. A simple SIS process: a susceptible calf (S) acquires infection either by the within-cohort transmission route (β) or the environmental transmission route (θ). An infected calf (I) can regain susceptible status again after recovery (γ).
Table 2. A summary of three properties that describe the datasets for both calf cohort studies
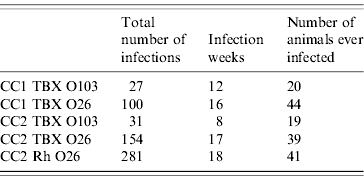
The identities of individual datasets and their abbreviations are as explained in the main text.
Modelling the sensitivity of agglutination test with TBX agar in detecting serogroup O26
In CC2, we observed that the agglutination test with TBX agar failed to detect almost half of O26-positive samples detected with rhamnose agar. If we ignore those seven TBX-positive rhamose-negative O26 samples from CC2, we then can estimate the relative sensitivity when using the TBX agar. Here, we define such a relative sensitivity as the probability of a sample testing positive when using TBX agar if it has already tested positive using rhamnose agar. Among the 281 samples which tested positive for serogroup O26 using rhamnose agar, 147 tested positive using TBX agar, giving a relative sensitivity for agglutination test with TBX agar of 0·53 (147/281). A new model (CC2 TBX O26S) can be constructed by incorporating this sensitivity into the basic model. We then fit this model to the dataset CC2 TBX O26 and determine its epidemiological parameters by using the same methodology as before. We can then compare the parameters estimated from our basic and sensitivity models for the dataset CC2 TBX O26 and investigate the effect of test sensitivity on parameter estimations.
PFGE type distribution
We extend the basic model to incorporate different PFGE types in order to explain the observed distribution of PFGE types in CC2 data. Models for PFGE types adopt the same basic model of transmission dynamics, cohort structure and sampling process as before. Whenever a calf is infected via the environmental transmission route, it then acquires a PFGE type randomly with equal probability from a total of M possible types in the environment. Here, we make a conservative assumption that the number of different PFGE types observed in the data reflects those present in the environment. Therefore, the number of different PFGE types in the environment, M, is 3 and 12 for serogroups O103 and O26 respectively. When the acquired infection is from the within-cohort route (i.e. from an infected calf), then the calf acquires the same PFGE type as the source. Finally, the assigned PFGE type is then lost when a recovery process occurs.
RESULTS
Best-fit parameter values
The best-fit parameter values for serogroups O26 and O103 in the two different calf cohort studies are summarized in Table 3. We illustrate our interpretation of these results for VTEC serogroup O103 in CC1, and the others can be interpreted in a similar manner. For serogroup O103 in CC1, the best parameter value for the within-cohort transmission coefficient (β) is 0·001/day per calf [95% confidence interval (CI) 0–0·008]: this implies that a susceptible calf acquires infection from a given infected calf on average every 1000 days (1/0·001). For the environmental transmission parameter (θ), the best value is 0·017/day (95% CI 0·009–0·035), i.e. a susceptible calf acquires infection from sources other than infected calves once on average every 59 days (1/0·017). The best-fit value for the recovery parameter (γ) is 0·38/day (95% CI 0·2–0·8), i.e. an infected calf remains infectious for an average of 2·6 days (1/0·38).
Table 3. A summary of model results: best-fit values of epidemiological parameters, the basic reproductive number, percentage of positive samples from within-cohort transmission route and the probability of consecutive infections due to persistence

Values in parentheses are the 95% confidence intervals for the appropriate model results. The identities of individual datasets and their abbreviations are as explained in the main text.
The basic reproductive number, R 0
With our parameter estimates, we can also calculate the basic reproductive number R 0, which is defined as the number of secondary infections produced from one primary infection:
where N is the population size, which equals 49 and 41 for CC1 and CC2 respectively. Table 3 summarizes values of R 0 for different models. Only the model fitted to the dataset CC2 TBX O103 has a value of R 0>1, others are all <1.
Sources of infection
Simulations of the models with best-fit parameters are run in order to determine the sources of individual infections. All of our models but one suggest that fewer than 20% of O26- or O103-positive samples are from the within-cohort transmission route (i.e. calf–calf transmission) whereas this percentage is 82% for VTEC O103 in CC2 (Table 3). Furthermore, it is also possible that none of the positive samples in the datasets CC1 TBX O103 and CC2 TBX O26 are from the within-cohort transmission route (95% CIs include zero). For each calf, we also keep a record of identities of two consecutive positive samples. Therefore the probability of one sample being positive as a result of persistence of the positive sample the week before can be calculated. Table 3 summarize those probabilities for all models. This probability is on average about 0·63, but it varies from as low as 0·39 to a much higher level of 0·85 among different datasets (Table 3).
Modelling the sensitivity of agglutination test with TBX agar in detecting serogroup O26
By using a test sensitivity value of 0·53, we fitted the sensitivity model (CC2 TBX O26S) to the dataset CC2 TBX O26 and estimated the three epidemiological parameters. The best-fit values for the within-cohort transmission coefficient (β), environmental transmission parameter (θ) and the recovery parameter (γ) are given in Table 3. These parameter estimates show reassuring consistency with those for CC2 Rh O26 (Table 3).
PFGE modelling
With equal probability of occurrence for all PFGE types in the environment, we simulated the PFGE models 10 000 times. For each realization, we calculated the variance of numbers of positive samples with particular PFGE types, and then ask where the variance calculated from the data is placed within the simulation distribution. For VTEC serogroup O103 detected with TBX agar, the data variance is 261·3, which is placed well within the 95% CI of the model variance (17·3–320·3). For VTEC serogroup O26 detected with TBX agar, the data variance is 56·4 and falls outside the 95% CI of the model variance (1·6–13·5). Similarly for VTEC serogroup O26 detected with rhamnose agar, the data variance (45·8) is not within the 95% CI of the data variance (1·7–12·0).
DISCUSSION
In this paper, we have investigated the epidemiology of VTEC serogroups O26 and O103 in two different calf cohorts on different farms. Both calf cohort studies have shown the environmental transmission parameter to be greater for serogroup O26 than for O103. This suggests that there may be a higher amount of VTEC O26 in the calves' environment than VTEC O103 on both farms. With the exception of serogroup O103 detected in CC2, the 95% CIs for the within-cohort transmission coefficient (β) do include zero, therefore evidence for transmission of VTEC between calves is not conclusive. Our results also suggest that, with the exception of VTEC O103 in CC2, fewer than 20% of O26- or O103-positive samples arise from the within-cohort transmission route. It has been recently suggested that VTEC can persist outside their hosts for an extended period of time in the order of weeks [Reference Duffy26], and VTEC are often found on farm surfaces [Reference Williams27]. Therefore, our findings on the majority of infections are from the environmental routes are probably of biologically relevance. Evidence for calf-to-calf transmission of VTEC O103 in the second calf cohort is stronger and by visual inspection of the data (Fig. 1) one can see that positive samples are clustered in the earlier stage of the study (in weeks 3 and 4). As for the recovery parameter (γ), our results suggest that VTEC O26- or O103-infected calves remain infectious for an average of 2·5 days during the first study, whereas this duration is approximately 5 days for CC2.
In a previous study Liu et al. [Reference Liu23] fitted similar mathematical models to data from the first calf cohort for 12 VTEC serogroups. In contrast to our approach here, where a model is fitted to an individual VTEC serogroup, Liu et al. [Reference Liu23] looked at all 12 VTEC serogroups collectively. All 12 serogroups were assumed to be governed by the same epidemiological process and a best-fit parameter set was estimated. The best-fit model incorporated heterogeneity in the recovery parameter, but all VTEC infections were estimated to be lost in less than 1 week, similar to the results reported here for O26 and O103 alone. Also confirmed by this study, the previous work suggested there were some calf-to-calf transmission of VTEC in the cohort but the majority of VTEC infections were from environmental sources.
From our estimates of epidemiological parameters, we can also determine the basic reproductive number (R 0) for VTEC serogroups O26 and O103 for the two different calf cohorts. Apart from serogroup O103 in CC2, all models have R 0 values of <1. This implies that VTEC are not able to successfully spread in the calf cohort without the reintroduction of VTEC from sources other than infected calves. However, one assumption in all of our models is that the environmental transmission parameter (θ) is constant throughout our studies. It is possible that the level of VTEC in the surrounding environment of a calf cohort might have spatial-temporal patterns resulting in clusters of VTEC outbreaks, in which case we would have over-estimated the within-cohort transmission coefficient (β) and the basic reproductive number (R 0).
In addition to TBX agar, rhamnose agar was also used to isolate VTEC serogroup O26 in CC2. Since rhamnose agar is known to have a good sensitivity and specificity in detecting VTEC O26 [Reference Hiramatsu21], this thus provides us an opportunity to check on what happens if we know that the isolation method with TBX agar is missing some O26-positive samples. Both TBX and rhamnose models produce consistent parameter estimate for the duration of infection. The major differences are in both transmission parameters with those for the rhamnose model being much higher than that for the TBX model. Furthermore, the basic reproductive number (R 0) for the rhamnose model is nearly ten times higher than that for the TBX model (although still <1). We have also constructed a model for serogroup O26-positive samples which allows for imperfect detection with TBX agar. This model gives excellent agreement with the fit to the rhamnose data for all three epidemiological parameters. Therefore, like other host–pathogen systems [Reference Coen28], our results suggest that test sensitivity is potentially a problem in characterizing the epidemiology of VTEC.
In CC2, all O103-positive samples and a subset of O26-positive samples had their PFGE patterns determined. Among O103 isolates, there was a single dominant PFGE type, while there were several abundant types among the O26 isolates. We were also interested in whether there were differences between different PFGE types in terms of their abundances in the environment surrounding the calf population. Our models suggest that a uniform distribution of different PFGE patterns in the environment can reproduce the observed heterogeneity in those O103 isolates. Assuming our finding of a higher calf-to-calf transmission rate for VTEC O103 is correct, then the observed heterogeneity in the PFGE patterns in CC2 could result from chance effects where one PFGE type was picked up by one calf followed by mini-outbreaks of this particular PFGE type. However, a uniform distribution of different PFGE types in the environment cannot explain the heterogeneity among O26 isolates. Assuming that our finding of little calf-to-calf transmission of VTEC O26 is correct, then the observed heterogeneity in PFGE distribution among O26 isolates might be a reflection of that in the environment.
This analysis is based on just two datasets and we suggest more longitudinal studies of the same kind are needed to test the robustness of our conclusions. Even so, a consistent picture is beginning to emerge from fitting simple mathematical models to longitudinal studies of VTEC infection in cohorts of calves. These infections are short-lived, typically lasting less than 1 week. Despite some variation between serotypes and between cohorts, VTEC are not usually sufficiently transmissible to persist in these cohorts without repeated introduction from some other source on the farm. These findings imply that levels of VTEC infection in young cattle can, in principle, be controlled by reducing contamination of their immediate environment.
ACKNOWLEDGEMENTS
This study was funded by the Wellcome Trust International Partnership Research Award in Veterinary Epidemiology (IPRAVE) project, and a Wellcome Trust funded Mathematical Biology Research Training fellowship to L. M. We thank Alastair W. Smith, Hazel I. Knight and Jude Evans for the processing of calf faecal samples during this study.
DECLARATION OF INTEREST
None.






