Salmonella enterica serotype Typhi (S. Typhi) is the causative agent of typhoid fever and is transmitted via food or water contaminated with human faeces. The bacterium is of great clinical importance as humans are the only recognized reservoir of S. Typhi and typhoid fever is a major cause of morbidity and mortality in humans, particularly in developing countries. For 2000, it was estimated that worldwide, typhoid fever caused 21 650 974 illnesses and 216 510 deaths [Reference Crump, Luby and Mintz1]. Antibiotics are critical in the management of typhoid fever. Various fluoroquinolones such as ciprofloxacin have become routine treatment for typhoid fever [Reference Aarestrup2]. However, worldwide, nalidixic acid-resistant [minimum inhibitory concentration (MIC) ⩾32 μg/ml] strains have emerged showing reduced susceptibilities to ciprofloxacin, and patients infected with such strains have been associated with ciprofloxacin treatment failures. Resistance to quinolones usually occurs as a result of alterations in the target enzymes (DNA gyrase and topoisomerase IV) and as a result of changes in drug entry and drug efflux [Reference Jacoby3]. Resistance to quinolones can also be mediated by plasmids which carry genes coding for Qnr proteins, which protect the quinolone targets from inhibition [Reference Jacoby, Chow and Waites4].
South Africa (SA) has an estimated burden of typhoid fever of 100 infections/100 000 of population per year [Reference Crump, Luby and Mintz1] and in eastern parts of the country there exist areas with a reservoir of typhoid fever. There are few published data on the occurrence of quinolone-resistant strains of S. Typhi in sub-Saharan Africa and SA although the occurrence of such strains has been reported in Kenya [Reference Kariuki5], Nigeria [Reference Akinyemi and Coker6] and Cameroon [Reference Nkemngu, Asonganyi and Njunda7]. In recent years, there has been a rapid increase in the prevalence of nalidixic acid resistance amongst S. enterica in SA. In the current study, we report on the prevalence of quinolone-resistant S. Typhi isolated from humans in SA over a 5-year period (2003–2007), molecular epidemiology analysis and probable mechanisms of quinolone resistance of these strains.
The Enteric Diseases Reference Unit (EDRU) of the National Institute for Communicable Diseases (NICD) is the national reference centre in SA for enteric pathogens. The Unit participates in national laboratory-based surveillance for human isolates of Salmonella and isolates are voluntarily submitted from more than 120 clinical microbiology laboratories across the country. For the current study, isolates were identified and serotyped using standard techniques. Susceptibility testing to antimicrobial agents was determined by the Etest (AB Biodisk, Sweden). Genotypic relatedness of isolates was investigated by pulsed-field gel electrophoresis (PFGE) analysis of XbaI-digested genomic DNA on a CHEF-DR III electrophoresis system (Bio-Rad Laboratories Inc., USA) using a PulseNet protocol. Isolates were also typed by multiple-locus variable-number tandem repeat analysis (MLVA) based on three variable-number tandem-repeat (VNTR) gene loci (TR1, TR2, TR3) as previously described [Reference Liu8] but modified to incorporate automated capillary electrophoresis of fluorescently labelled PCR products. Electrophoresis of PCR products was performed through POP-7 polymer (Applied Biosystems, USA) at 15 kV for 25 min at 60°C using an Applied Biosystems 3130 genetic analyser. Raw data was analysed using GeneMapper (version 4.0) software (Applied Biosystems) which identified each VNTR locus and sized the gene product by comparison with an internal size standard. PCR was used to isolate the quinolone resistance-determining region (QRDR) of gyrA, gyrB, parC and parE from isolates using previously described methods [Reference Eaves9]. Genes were sequenced using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems) and an Applied Biosystems 3130 genetic analyser; sequences were analysed for mutations using DNASTAR Lasergene (version 8.0) software (DNASTAR Inc., USA). PCR was used to test for the presence of qnrA, qnrB and qnrS genes in isolates using previously described methods [Reference Jacoby, Chow and Waites4, Reference Jacoby10, Reference Hopkins11]. Efflux of quinolones out of bacterial cells was investigated as follows. For nalidixic acid and ciprofloxacin, agar dilution MIC testing of isolates was performed in the absence and presence of 40 μg/ml of the efflux pump inhibitor (EPI), Phe-Arg-β-naphthylamide [Reference Baucheron12]. MIC testing was performed according to the methods of the Clinical and Laboratory Standards Institute.
For the years 2003–2007, the EDRU received 8464 human isolates of Salmonella, 510 (6%) of which were S. Typhi; 27 proved to be nalidixic acid-resistant. These were distributed over the study period as follows: 2003 (3), 2004 (5), 2005 (11), 2006 (4) and 2007 (4). The resistant isolates showed nalidixic acid MICs of 128–512 μg/ml and also reduced susceptibility to ciprofloxacin (MICs 0·125–0·5 μg/ml). Only 19 isolates remained viable for further study and these were differentiated by PFGE into five DNA pattern types (Table 1). However, MLVA was more discriminatory and distinguished 10 types (Table 1) particularly in the most frequent PFGE type 2 cluster (14 isolates) where six MLVA types were identified. This relatively high level of genetic diversity suggests that resistant strains usually emerged independently of one another.
Table 1. Summary of data for 19 quinolone-resistant Salmonella Typhi isolates from South Africa
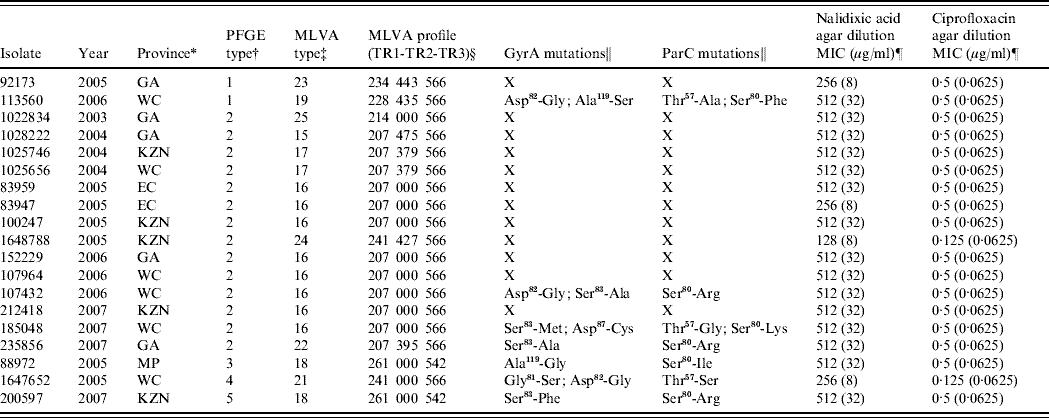
PFGE, Pulsed-field gel electrophoresis; MVLA, multiple-locus variable-number tandem repeat analysis; MIC, minimum inhibitory concentration.
* GA, Gauteng; WC, Western Cape; KZN, KwaZulu-Natal; EC, Eastern Cape; MP, Mpumalanga.
† PFGE patterns were analysed using the BioNumerics (version 5.1) software (Applied Maths, Belgium). Dendrograms of the patterns were created using the unweighted pair-group method with arithmetic averages, with analysis of patterns incorporating the dice-coefficient at an optimization setting of 0·5% and a position tolerance setting of 1·5%. A PFGE type was defined as a group of patterns sharing ⩾90% similarity on dendrogram.
‡ A MLVA type was defined by a unique MLVA profile.
§ 000 represents no amplification of the PCR product at locus TR2.
‖ X, isolates not analysed for mutations.
¶ The value in parentheses indicates the result of MIC testing within the presence of an efflux pump inhibitor.
A 16- to 32-fold decrease in nalidixic acid MIC and a 2- to 8-fold decrease in ciprofloxacin MIC in the presence of EPI was observed for the resistant isolates (Table 1), suggesting that efflux of antibiotic out of bacterial cells contributes to the development of quinolone resistance. Efflux pump-mediated quinolone resistance is well described in Salmonella, of which the AcrAB-TolC efflux system plays a major role [Reference Baucheron13]. This system is inhibited by the EPI compound used in this study indicating that this same system is operative in our quinolone-resistant isolates.
As most of our isolates still showed nalidixic acid resistance with MICs at 32 μg/ml in the presence of an EPI (Table 1), we proceeded to identify additional mechanisms of resistance. PCR screening for the three major qnr families (qnrA, qnrB, qnrS) was negative, thus excluding plasmid-mediated Qnr proteins as a mechanism of resistance. Seven resistant isolates were examined for mutations in the QRDR of gyrA, gyrB, parC and parE. As expected, no amino-acid mutations were identified in GyrB and ParE. However, all seven isolates showed amino-acid mutations in both GyrA and ParC (Table 1). For quinolone-resistant Salmonella, GyrA mutations mostly occur at positions Ser83 and Asp87 [Reference Eaves9, Reference Ling14]. Most isolates similarly showed mutations at Ser83 (to Ala, Met or Phe) or Asp87 (to Cys), with isolate 185 048 also showing a double mutation at positions 83 and 87. However, some of the isolates lacked mutations at positions 83 and 87 and instead they showed the following mutations in GyrA; isolate 88 972 showed an Ala119 (to Gly) mutation, while isolate 113 560 showed Asp82 (to Gly) and Ala119 (to Ser) mutations. The commonality between these two isolates was the GyrA mutation at position 119, so we hypothesize that this position may also be important for development of quinolone resistance in Salmonella. Mutation at position 119 is not commonly reported, although it previously has been identified in nalidixic acid-resistant S. Typhimurium [Reference Griggs, Gensberg and Piddock15]. For quinolone-resistant Salmonella, ParC mutations mostly occur at positions Thr57, Thr66 and Ser80 [Reference Eaves9, Reference Ling14]. Our isolates similarly showed mutations at Thr57 (to Ala, Gly, or Ser) or Ser80 (to Phe, Arg, Lys or Ile); 113 560 and 185 048 also showed a double mutation at positions 57 and 80. Analysis of the GyrA and ParC mutation data has predicted that isolate 185 048, with a double mutation in GyrA (positions 83 and 87) in combination with a ParC mutation at position 80, should be resistant to ciprofloxacin; however, it remains susceptible (MIC 0·5 μg/ml). This combination of mutations is well described as being associated with ciprofloxacin resistance [Reference Ling14]. Isolate 185 048 also showed a Thr57 (to Gly) mutation in ParC, which may convey ciprofloxacin susceptibility on the isolate. Eaves et al. [Reference Eaves9] suggested that a ParC mutation at position 57 is a naturally occurring compensatory mutation that makes isolates more sensitive to ciprofloxacin but not to nalidixic acid. Our data support this.
In conclusion, the prevalence of nalidixic acid resistance in isolates of S. Typhi in SA for the years 2003–2007 was around 5%. Nalidixic acid-resistant isolates were genetically diverse which suggested that resistant isolates usually emerged independently of one another. The mechanism of resistance is most likely amino-acid mutations in GyrA and ParC in combination with active efflux of antibiotic out of the bacterial cell.
ACKNOWLEDGEMENTS
This work was financially supported by the following South African institutions: the National Health Laboratory Service and the Medical Research Council.
DECLARATION OF INTEREST
None.



