Introduction
Viral hepatitis refers to a group of infectious disorders caused by various kinds of hepatitis viruses (hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV) and hepatitis E virus (HEV)), and it could lead to life-threatening conditions including cirrhosis, liver failure or hepatocellular carcinoma [Reference Lanini1, Reference Asrani2]. Although the exact mechanism of its pathogenesis is still uncertain, it was believed that genetic architecture was essential for the development of viral hepatitis. In the first place, the incidences of viral hepatitis in different populations vary greatly [Reference Jefferies3, Reference Pardee4], and genetic background was probably one of the reasons behind differences in disease prevalence across different populations. In the second place, previous association studies also identified numerous susceptible genetic loci of viral hepatitis [Reference Karlsen, Lammert and Thompson5, Reference Lammert6]. However, genetic factors that contribute to the development of viral hepatitis are still not fully elucidated.
Cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) and interleukin-18 (IL-18) are pro-inflammatory cytokines, and they both serve as crucial modulators of anti-viral immune responses [Reference Chikuma7, Reference Kaplanski8]. Therefore, if a genetic polymorphism could alter the transcription activity of CTLA-4/IL-18 or the protein structure of CTLA-4/IL-18, it is biologically plausible that this polymorphism may also impact anti-viral immune responses and confer susceptibility to many types of infectious diseases including viral hepatitis.
In the past 20 years, results about associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis were already reported by many association studies, yet the conclusions of these studies were still inconsistent. To better analyse associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis, we carried out this study to get a more statistically reliable conclusion by combing the results of all relevant studies.
Materials and methods
This meta-analysis was written in accordance with the PRISMA guideline [Reference Moher9].
Literature search and inclusion criteria
To retrieve eligible articles, we searched Pubmed, Web of Science and Embase using key words listed below: (‘interleukin-18’ or ‘IL-18’ or ‘interleukin 18’ or ‘IL 18’ or ‘cytotoxic T lymphocyte antigen-4’ or ‘CTLA-4’) and (‘polymorphism’ or ‘variant’ or ‘variation’ or ‘mutation’ or ‘SNP’ or ‘genome-wide association study’ or ‘genetic association study’ or ‘genotype’ or ‘allele’) and (‘viral hepatitis’ or ‘chronic hepatitis’ or ‘acute hepatitis’ or ‘Hepatitis A’ or ‘Hepatitis B’ or ‘Hepatitis C’ or ‘Hepatitis D’ or ‘Hepatitis E’ or ‘HAV’ or ‘HBV’ or ‘HCV’ or ‘HDV’ or ‘HEV’). The references of retrieved articles were also screened by us in case some related articles may be missed by our electronic literature searching.
To be included in this meta-analysis, some criteria must be satisfied: (I) studies about associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis in humans; (II) offer genotypic or allelic distribution of CTLA-4/IL-18 polymorphisms in patients with viral hepatitis and controls and (III) full manuscript in English or Chinese is retrievable. We only included the most up-to-date study if duplicate reports were found during literature search.
Data extraction and quality assessment
Two authors extracted following information from eligible articles: (I) name of the first author; (II) published year; (III) country where the study was conducted; (IV) ethnicity of involved participants; (V) number of patients with viral hepatitis and controls in each study and (VI) genotype distributions of polymorphisms in CTLA-4/IL-18 among patients with viral hepatitis and controls. P Values of Hardy–Weinberg equilibrium (HWE) were also calculated.
The authors used Newcastle–Ottawa scale (NOS) to assess the methodology quality of eligible articles [Reference Stang10]. The score range of NOS is between zero and nine, when an article got a score of seven or more, we considered that the methodology of this publication was good.
Two authors extracted data and assessed the quality of eligible articles. The authors wrote to the corresponding authors for additional information if essential information was found to be incomplete.
Statistical analyses
We used Review Manager to combine the results of individual studies. The Z test was employed to assess associations between polymorphisms in CTLA-4/IL-18 and susceptibility to viral hepatitis in dominant, recessive, over-dominant and allele comparisons. All CTLA-4/IL-18 polymorphisms contain a major allele (M) and a minor allele (m), the dominant comparison is defined as MM vs. Mm + mm, recessive comparison is defined as mm vs. MM + Mm, over-dominant comparison is defined as Mm vs. MM + mm and the allele comparison is defined as M vs. m. The statistical significant threshold of the P value was set at 0.05. I 2 statistics were used to assess between-study heterogeneities. Random-effect models (DerSimonian–Laird method) were used to combine the results if I 2 is larger than 50%. Otherwise, fixed-effect models (Mantel–Haenszel method) were used to combine the results. We also carried out subgroup analyses first by type of disease and then by ethnicity of participants. We examined the stability of combined results by deleting one study each time and combining the results of the remaining studies. Funnel plots were used to estimate whether our combined results may be influenced by overt publication biases.
Results
Characteristics of included studies
We identified 271 articles during literature searching. Fifty-nine articles were assessed for eligibility after excluding unrelated or duplicate publications. We further excluded 16 reviews and four case controls, and another two articles were excluded because of missing crucial data. Totally 37 articles were ultimately included in this meta-analysis (Fig. 1). Extracted data of eligible articles are shown in Table 1.

Fig. 1. Flowchart of study selection for the present study.
Table 1. The characteristics of included studies for this meta-analysis

CTLA-4, cytotoxic T-lymphocyte-associated antigen 4; IL-6, interleukin-6; HBV, hepatitis B virus infection; HCV, hepatitis C virus infection; HWE, Hardy–Weinberg equilibrium; NOS, Newcastle–Ottawa scale; NA, not available.
Meta-analysis results for polymorphisms in CTLA-4 and viral hepatitis
Fourteen eligible articles were about CTLA-4 polymorphisms and viral hepatitis. CTLA-4 rs231775 (recessive comparison: OR 1.33, 95% CI 1.23–1.43) polymorphism was found to be significantly associated with viral hepatitis in overall combined analyses. Further subgroup analyses revealed similar positive findings for CTLA-4 rs231775 polymorphism in HBV, especially among East Asians. A significant association with HCV was also detected for CTLA-4 rs5742909 polymorphism (see Table 2).
Table 2. Meta-analysis results of this study
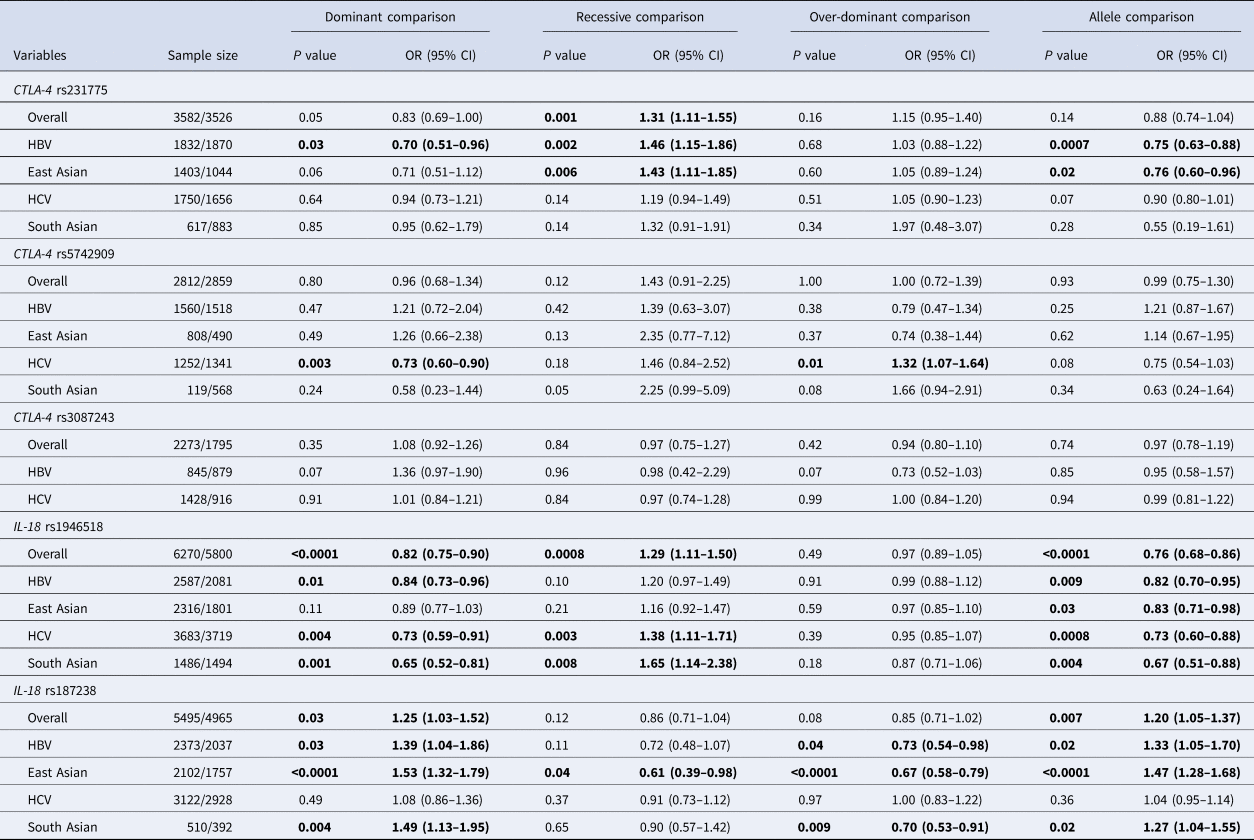
OR, odds ratio; CI, confidence interval; NA, not available; HBV, hepatitis B virus infection; HCV, hepatitis C virus infection.
The values in bold represent there is statistically significant differences between cases and controls.
Meta-analysis results for polymorphisms in IL-18 and viral hepatitis
Twenty-three articles were about IL-18 polymorphisms and viral hepatitis. IL-18 rs1946518 (dominant comparison: OR 0.82, 95% CI 0.75–0.90; recessive comparison: OR 1.29, 95% CI 1.11–1.50; allele comparison: OR 0.76, 95% CI 0.68–0.86) and IL-18 rs187238 (dominant comparison: OR 1.25, 95% CI 1.03–1.52; allele comparison: OR 1.20, 95% CI 1.05–1.37) polymorphisms were both found to be significantly associated with viral hepatitis in overall combined analyses. Further subgroup analyses revealed similar positive findings for IL-18 rs1946518 and rs187238 polymorphisms in HBV, especially among East Asians. Moreover, we also found that IL-18 rs1946518 and rs187238 polymorphisms were significantly associated with susceptibility to HCV, especially among South Asians (see Table 2).
Sensitivity analyses
Stabilities of combined results were examined by deleting one study each time and combining the results of the remaining studies. The trends of associations remained consistent in sensitivity analyses, indicating that the combined results were statistically stable.
Publication biases
Funnels plots were employed to estimate whether our combined results may be influenced by overt publication biases. Funnel plots were overall symmetrical, indicating that the combined results were unlikely to be seriously impacted by overt publication biases (see Supplementary Fig. S1).
Discussion
The combined results of this meta-analysis revealed that CTLA-4 rs231775, IL-18 rs1946518 and IL-18 rs187238 polymorphisms were significantly associated with susceptibility to HBV, especially among East Asians. Moreover, CTLA-4 rs5742909, IL-18 rs1946518 and IL-18 rs187238 polymorphisms were also found to be significantly associated with susceptibility to HCV, especially among South Asians. The trends of associations remained consistent in sensitivity analyses, indicating that the combined results were stable.
Some points need to be considered when interpreting our findings. First, past pre-clinical studies found that rs231775, rs5742909 and rs3087243 polymorphisms in CTLA-4 as well as rs1946518 and rs187238 polymorphisms in IL-18 could alter transcription activity or protein structure of CTLA-4/IL-18 [Reference Gough, Walker and Sansom11–Reference Sedimbi, Hägglöf and Karlsson14]. So these variations may influence biological function of CTLA-4/IL-18, result in immune dysfunction, impact anti-viral immune responses and ultimately confer susceptibility to viral hepatitis. Thus, our meta-analysis may be statistically insufficient to observe the real underlying associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis in certain groups. Therefore, future studies with larger sample sizes still need to confirm our findings. Second, according to our searching results, studies about HBV were mainly conducted in East Asians, whereas studies in HCV were mainly conducted in South Asians. So we call on scholars to examine associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis in other populations. Third, the etiologies of viral hepatitis are extremely complex, so we highly recommend further genetic association studies to explore the effects of haplotypes and gene–gene interactions on disease susceptibility [Reference Nishi15]. Fourth, we aimed to investigate associations between all polymorphisms in CTLA-4/IL-18 and viral hepatitis in the very beginning. However, we did not find any study on other CTLA-4/IL-18 polymorphisms. Nor did we find any studies about HAV, HDV or HEV. So we only focused on associations of five polymorphisms with HBV and HCV in this meta-analysis.
Like all meta-analyses, this study also has some limitations. Firstly, the results regarding associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis were based on combining unadjusted findings of eligible studies due to the lack of raw data [Reference He16]. Secondly, the relationship between polymorphisms in CTLA-4/IL-18 and viral hepatitis may also be affected by environmental factors. Nevertheless, the majority of eligible studies only focused on associations between polymorphisms in CTLA-4/IL-18 and viral hepatitis, so we could not explore genetic-environmental interactions in this meta-analysis [Reference Stättermayer and Ferenci17]. Thirdly, grey literatures were not searched. Thus, despite that funnel plots were overall symmetrical, we still could not rule out the possibility that our combined results may be affected by potential publication biases [Reference Moudi, Heidari and Mahmoudzadeh-Sagheb18].
Conclusions
In summary, this meta-analysis demonstrated that CTLA-4 rs231775, IL-18 rs1946518 and IL-18 rs187238 polymorphisms may confer susceptibility to HBV in East Asians, while CTLA-4 rs5742909, IL-18 rs1946518 and IL-18 rs187238 polymorphisms may confer susceptibility to HCV in South Asians. However, it should be noted that the combined results of this meta-analysis should still be confirmed by future studies with larger sample sizes.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268819001997.
Acknowledgements
None.
Author contributions
Yang Yu, Jie Qu and Guangqiang You conceived and designed the study. Yang Yu and Jie Qu conducted the literature review. Chen Zhou analysed data. Yang Yu, Jie Qu and Guangqiang You drafted the manuscript. All authors have read and approved the final manuscript.
Financial support
None.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors, thus ethical approval and informed consent are not required.
Data
Data sharing is not applicable to this article as no new data were created or analysed in this study.





