Introduction
Shiga toxin-producing Escherichia coli (STEC) are a group of bacteria associated with human disease and are defined by the presence of one or both phage encoded Shiga toxin genes; stx1 and stx2. Compared to other bacterial pathogens, it is a relatively rare infection in many parts of the world but is of public health concern due its low infectious dose (<100 bacteria) [Reference Byrne1] and potential to cause severe disease (3).
Worldwide, it is estimated that there are around 2.8 million cases annually, leading to 3890 cases of haemolytic uraemic syndrome (HUS) and 230 deaths [Reference Majowicz2]. The Europe wide rate of infection is estimated to be 1.4 cases per 100 000 population but reported rates vary between countries (range: <0.1–12.4 cases per 100 000 population) [3]. The O157 STEC serogroup is most commonly associated with human disease in the United Kingdom; however, other serogroups are seen more frequently in other European countries [3]. Rates of infection in England have remained fairly constant for many years [Reference Adams4]. Europe has shown a similar pattern with an increase since 2011 attributed to wider use of molecular methods following a large outbreak linked to sprouted fenugreek seeds [Reference Buchholz5]. Rates of infection are highest in children and most cases occur in the late summer, at least in temperate areas, and this pattern is seen universally [Reference Lal6].
Healthy cattle are the main reservoir of STEC although they are also carried by sheep and other animals [Reference Persad and LeJeune7]. Animals shed a range of phage types (PTs) with the most prevalent in UK cattle being PT21/28, 8 and 34, PT 4 in sheep and PT2 in pigs [Reference Chapman8, Reference Milnes9]. STEC O157 survives well in the environment, remaining viable for many months in temperate conditions [Reference Avery, Moore and Hutchison10, Reference Williams11].
Transmission to humans occurs through multiple routes. Cases present themselves sporadically (occurring independently of other cases) or as part of small outbreaks due to person-to-person spread in closed settings, particularly childcare facilities. The low infectious dose of STEC means that once in the population, person-to-person spread is common [Reference Byrne1]. Larger outbreaks tend to be associated with foodborne transmission, with an increasing trend towards salad vegetables and away from meat and dairy products [Reference Adams4]. Direct contact with the farming environment or ruminants, such as in open farms or petting zoos [Reference Byrne1], is an important risk factor for STEC infection [Reference O'Brien, Adak and Gilham12]. Indirect contact with animals or environments contaminated with their faeces is also of importance but the actual process leading to infection is less well understood. Heavy rainfall and flooding events can lead to contamination of fresh and marine water systems [Reference Williams13], beaches [14–Reference Ihekweazu18] and food crops. Poorly managed private water supplies (PWS) present a particular risk in areas of high animal density [Reference Richardson19, Reference Risebro, Pitchers and Hunter20].
Phylogenetic analysis of strains circulating in humans and UK cattle during 2014 described three distinct lineages (I, II and I/II) descended from a common ancestor. Lineage I contains PT 21/28 and PT32; strains encoding stx2 only and associated with more severe disease. Lineage II contains PT8 and Lineage I/II PT2. Isolates from humans and UK cattle are closely related suggesting that PT8 and, in particular, PT21/28, have a domestic source and are domestically acquired [Reference Dallman21]. With the advent of routine whole genome sequencing (WGS), it is now possible to identify links between cases that previously appeared sporadic in nature. These cases may exhibit spatial clustering, sometimes over long periods of time, suggesting that geographically restricted transmission of highly related strains can occur [Reference Butcher22].
Ecological studies in the United Kingdom, Europe and further afield demonstrate a spatial association between rates of infection, cattle density and other factors and all describe a seasonally driven picture with rates highest in the summer [Reference Innocent23–Reference Ohaiseadha29]. There are limitations to these studies. Sheep were only considered in one study [Reference Ohaiseadha29], despite being a known reservoir. The study populations were restricted to children [Reference Haus-Cheymol25] or focussed upon severe disease [Reference Haus-Cheymol25]. Some studies may also have included cases linked to outbreaks, a potential source of bias which are not ideal as the source of their infection may have differed from sporadic cases [Reference Pearl28]. Cases reporting foreign travel were included in some studies [Reference Haus-Cheymol25, Reference Frank26, Reference Pearl28], but not others [Reference Innocent23–Reference Haus-Cheymol25]. Finally, all of these studies were performed at serogroup level only, even though different subgroups may have different sources and hence potentially different risk factors.
In this study we overcome these limitations using enhanced surveillance of STEC which has been performed in England since 2009. These data arguably represent the most comprehensive data set for STEC infections in the world. This allows accurate identification of cases who have been part of an outbreak (and so are not representative of all cases) and those who report travel abroad or within the United Kingdom
This study had three aims. The first was to describe the spatial and temporal distribution of sporadic STEC O157 cases in England, the second was to test the relationship between the numbers of infections and hypothesised risk factors and the third was to test whether these risks differed by STEC subtype. Finally we explored how these risks varied between all sporadic cases and sporadic cases when those reporting national or foreign travel are excluded.
Methods
Isolates of E. coli O157 identified locally are sent for confirmation and typing at the Gastrointestinal Bacterial Reference Unit (GBRU). Detection and confirmation of STEC includes biochemical identification and serotyping of bacterial isolates. Since 1989, strains belonging to E. coli O157 have been further differentiated by using a phage-typing scheme developed in Canada [Reference Adams4].
The National Enhanced Surveillance System for STEC (NESSS) was introduced in England in 2009. The system collects clinical and epidemiological information for each laboratory confirmed case using a standardised questionnaire. This information is linked to reference microbiology information including PT, presence of virulence factors and whole genome sequence data.
The case definition for the purpose of this study was a sporadic case of STEC O157, confirmed by GBRU and reported to NESSS between 1 January 2009 and 31 December 2015. An overview of the data selection process is shown in Figure 1.
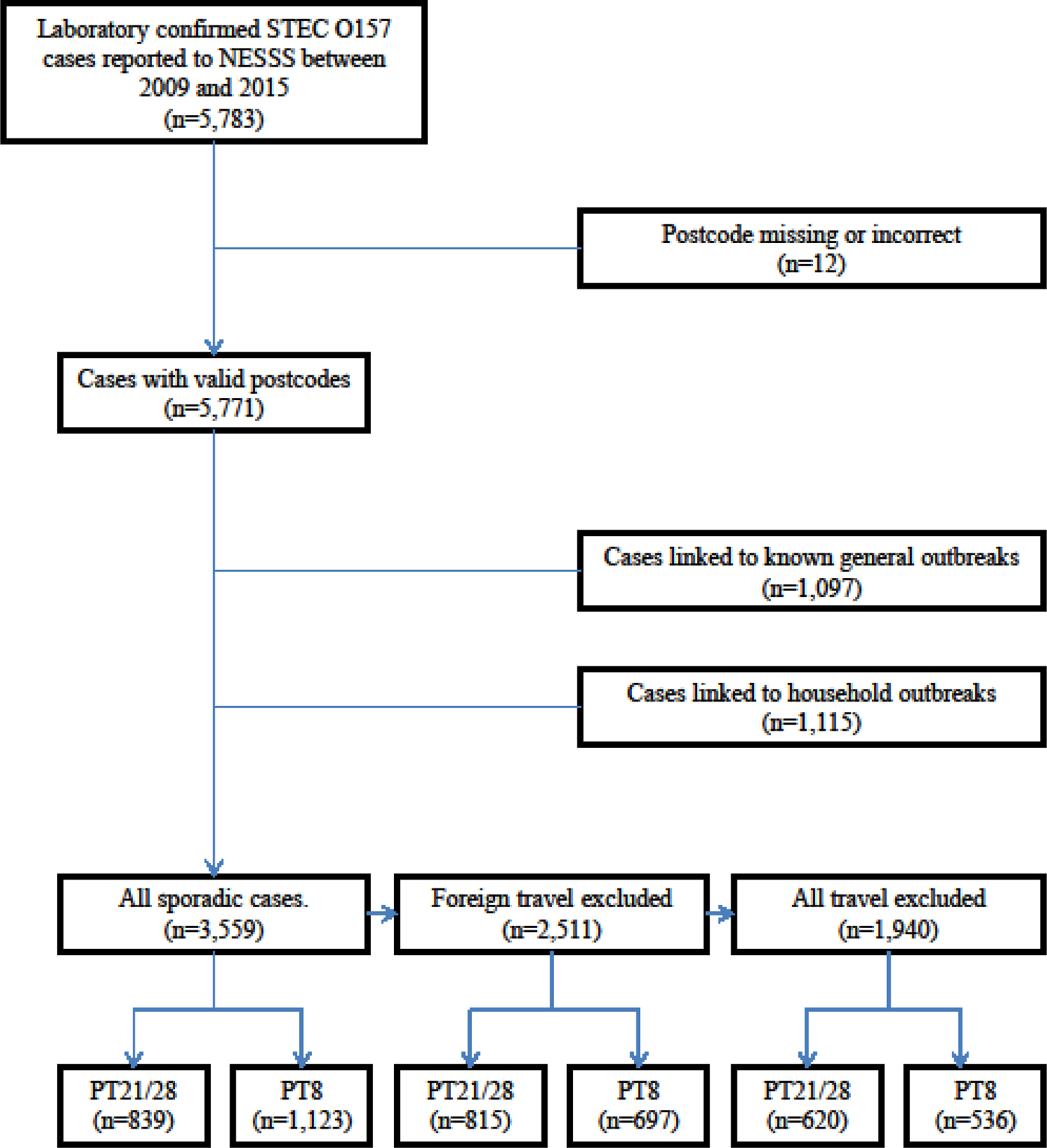
Fig. 1. Flow chart for case selection.
The main aim of our analysis was to estimate the effect of hypothesised risk factors on the occurrence of sporadic cases (i.e. those occurring independently of each other). We therefore excluded cases linked to known outbreaks because their residential location rarely reflects exposure to the source of their infection, particularly for large outbreaks linked to widely distributed foodstuffs. Cases linked to household outbreaks were also excluded. Household outbreaks were identified as those where at least two cases had isolates of the same serotype and PT that were collected within 6 months of each other, processed by a laboratory in the same Health Protection Team area and sharing the same surname and/or UK postcode.
The postcode (an alphanumeric reference developed by the UK Post Office to facilitate the delivery of mail and each containing around 15 addresses) for each case was geocoded to provide a spatial reference, allow visual display of the location of cases and enable the details of each case to be spatially joined to other data sets at the lower super output area (LSOA) level defined by the Office for National Statistics (ONS) [30]. LSOAs were chosen because they provide the most homogenous unit in terms of geographical size (mean 3.9 km2, range 0.02–684 km2) and population size (mean 1613 persons, range 985–8300 persons).
Crude incidence rates were calculated using the total population denominator data for each LSOA drawn from the last ONS Census performed in 2011.
For each LSOA, a dependent variable, indicating the number of cases that occurred during the study period was created. Because STEC is an uncommon infection, the majority (90.1%) of LSOAs had no cases, 9.3% had one case and 0.6% had more than one case.
Dependent variables were created for all PTs, PT21/28 and PT8, further divided into three classes (all reported cases, those not reporting foreign travel and those reporting no travel either within the United Kingdom or abroad) giving a total of nine dependent variables.
The following explanatory variables were constructed for each LSOA:
Livestock density variables for cattle, sheep and pigs were calculated using the agricultural census of 2010 [31]. This census is performed every 10 years by the Department for the Environment, Food and Rural Affairs and collects detailed information on land usage and livestock populations. Farm level data are aggregated to a 5 × 5 km grid and individual farms are not identified.
Estimates of deprivation were obtained from the ONS. The index of multiple deprivation (IMD) was obtained from England in 2011 [32], which provides a set of relative measures of deprivation for LSOAs. This is based on seven domains of deprivation (income, employment, education, health, crime, housing and living environment). These domains are combined and ranked to produce the overall IMD score for each LSOA. For our analysis these data were divided into quintiles where quintile 1 is most deprived.
The degree of rurality for each LSOA was derived from the ONS rural urban classification used to distinguish rural and urban areas in England and Wales in 2011[Reference Bibby and Brindley33]. The classification defines areas as rural if they are outside settlements with more than 10 000 resident population. For LSOAs, there are four urban classes (major conurbations, minor conurbations, cities and towns, cities and towns in sparse settings) and four rural classes (town and fringe, town and fringe in a sparse setting, village and dispersed settlements, village and dispersed settlements in sparse settings). Due to the small numbers of cases resident in areas considered sparse, we grouped these eight classes into five by merging the three sparse categories with the corresponding non-sparse categories.
Outbreaks have been linked to beaches [14, Reference Ihekweazu18], hence the straight-line distance from the centroid of each LSOA to the GB coastline was calculated in kilometres.
Inland water coverage was identified as a risk factor in a Finnish study [Reference Jalava27]. The shapes and areas of inland water features were extracted from the Ordnance Survey MasterMap Topography Layer [34], summed and divided by the area of each LSOA to provide a proportional measure of fresh water coverage.
For each LSOA, the count of PWS was calculated using data submitted by local authorities to the Drinking Water Inspectorate during 2016. Local authorities are responsible for the enforcement and monitoring of PWS Regulations 2005 which require PWS to meet certain standards and for the location of each supply to be recorded. Three classes were created (0, 1–20 and >20 supplies).
Because the data sets used for inland water coverage and animal density differed from LSOAs in terms of geographical area or shape, we used a geographical information system (GIS) overlay function to create proportional measure in kilometre for each LSOA.
We used Jenks’ natural breaks method to create four categorical variables for each animal species, distance from the coast and inland water coverage. This method is designed to determine the best arrangement of values into different classes by seeking to minimise the variance within classes and maximise the variance between classes [Reference Jenks and Caspall35].
Statistical analysis
We considered three methods of regression analysis: Poisson, negative binomial and zero-inflated Poisson. The results of a likelihood ratio test of alpha and goodness of fit test following Poisson regression indicated that the data were overdispersed. The same analysis was repeated using negative binomial and zero-inflated negative binomial regression respectively. The Vuong test was not significant indicating that the standard negative binomial approach was best suited to the data. Proceeding with the negative binomial regression approach we conducted a multivariable analysis for each dependent and the independent variables. The first set of dependent variables were all sporadic cases, all sporadic cases minus those reporting foreign travel and all sporadic cases with no foreign or domestic travel. Two further models were then produced focussing upon PT21/28 and PT8 only. Person years (the total population of each LSOA multiplied by the years of observation) were included as an exposure variable. None of the multivariable analyses showed any associations with the distance from the coast and inland water coverage variable; hence these were removed from the analysis. The remaining independent variables were all included in the nine models to allow greater comparability between models. The dependent and independent variables were checked for correlation using Spearman's rank test. All coefficients showed low-to-moderate correlation with the exception of cattle and sheep density with a coefficient of 0.7. An analysis for collinearity indicated that the addition of each independent variable in turn did not lead to significant changes in the coefficients or significance of any other variables in the model. Presenting each livestock density variable to the model as continuous variables did not affect the results.
Results are presented in terms of incidence rate ratio (IRR) estimates and the 95% confidence interval (CI). The overall significance for a variable was estimated using the Wald test. All statistical analyses were performed using Stata version 13 [36].
Results
Rates of infection
A total of 3559 (34%) cases were eligible for inclusion in the statistical analysis (Fig. 1).
The crude incidence of all sporadic confirmed STEC O157 cases (including those reporting foreign travel) reported during the study period was 9.1 per million person years. The rural rate (13.3 per million person years) was 1.6 times higher than that of the urban rate (8.1 per million person years). Rates varied across the country with the highest in the North of the country, the North West, Midlands and the South West Peninsula (Fig. 2a) and this was seen each year during the study (Fig. 3).
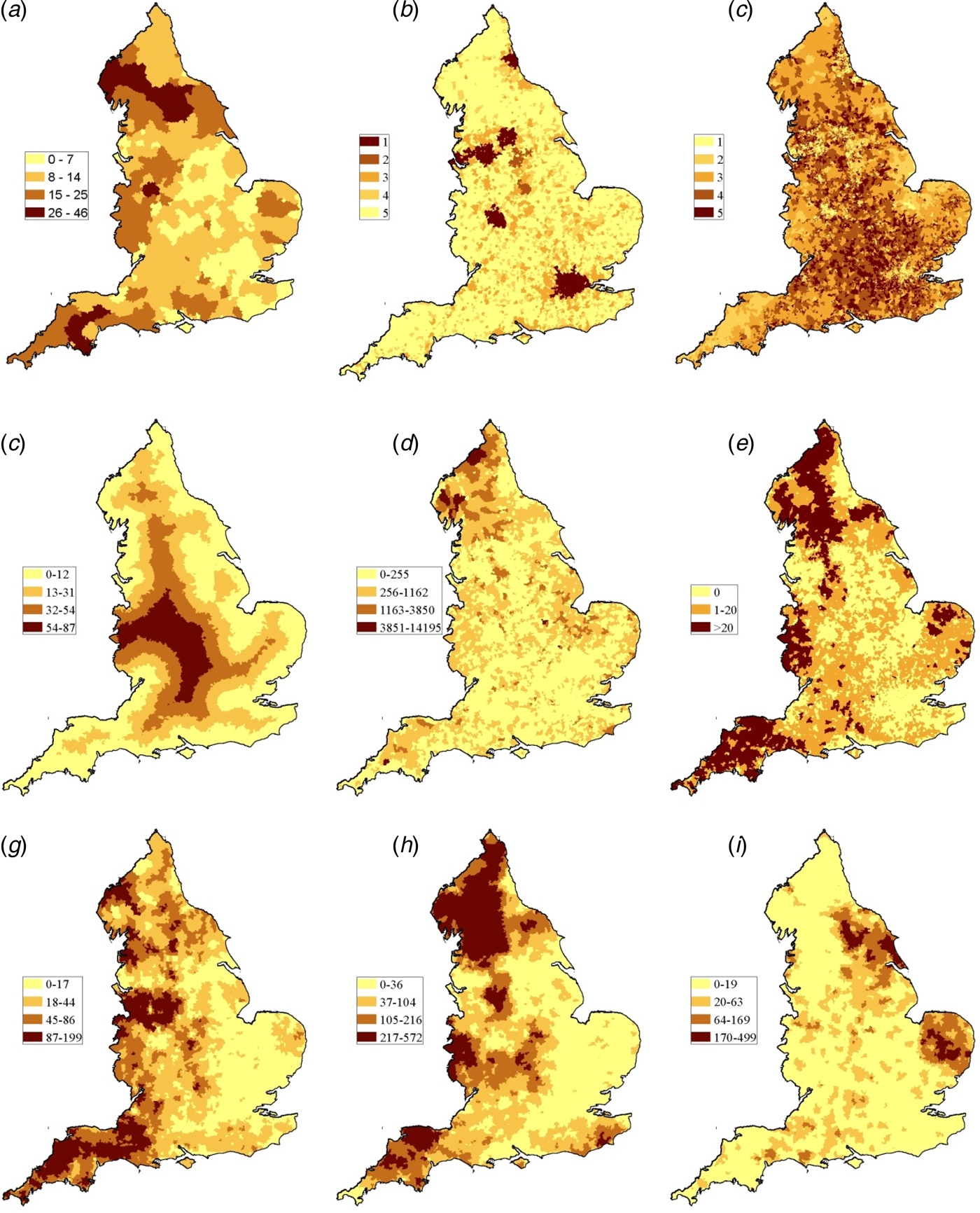
Fig. 2. (a) Cumulative incidence rate (sporadic cases/million person years) and spatial distribution of the eight independent variables used in the analysis (b) Residence (1: urban–major conurbation, 2: urban–minor conurbation, 3: urban–city and town, 4: rural–town and fringe, 5: rural-village. (c) IMD (deciles). (d) Distance from LSOA centroid to GB coast in kilometre. (e) Proportion of inland fresh water coverage of each LSOA (km2). (f) Numbers of PWS in each LSOA. (g) Cattle density (animals/km2). (h) Sheep density (animals/km2). (i) Pig density (animals/km2).
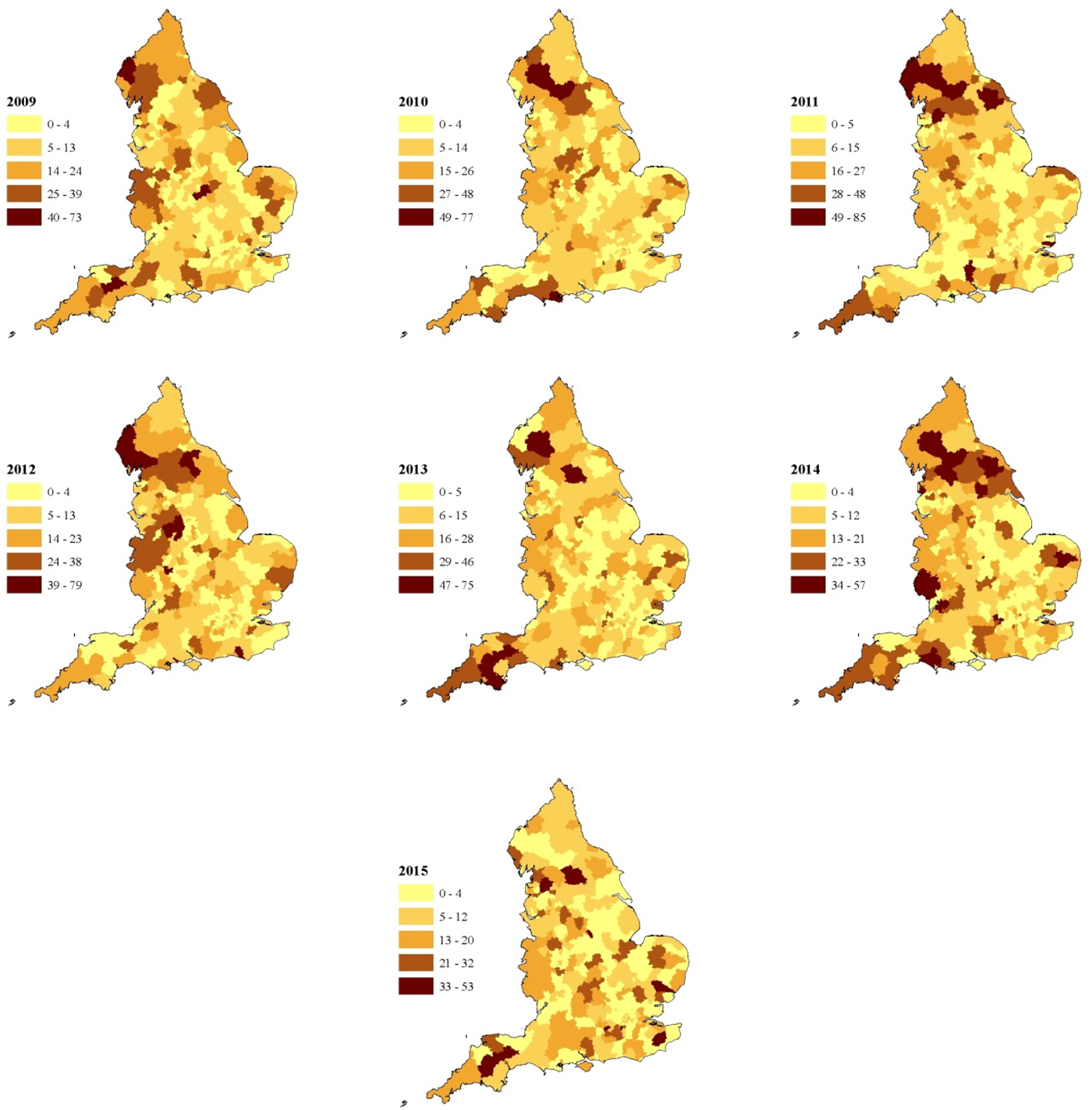
Fig. 3. Annual incidence rate of STEC O157 per million population including cases reporting travel outside the United Kingdom in England between 2009 and 2015 (unit of analysis is a local authority area).
The crude incidence rate of PT21/28 was 2.5 per million person years and for PT8 it was 3.3 per million person years. There was a distinct seasonality both in rural and urban areas with rates comparable during the winter but higher in rural areas during the summer (Fig. 4). The rate of infection declined from 2012, particularly for PT21/28 infections in rural areas (Fig. 4).

Fig. 4. Monthly rate of sporadic STEC O157 infection per million population in urban and rural settings in England between 2009 and 2015 (travel included).
The spatial distribution of animals varied across the country (Fig. 2g–i). The mean cattle density ranged from 0 to 199 animals/km2 with the highest densities observed in the South West Peninsula, areas of the North West (Cheshire) and Midlands (Staffordshire) and in the North. Sheep density ranged from 0 to 572 animals/km2 with the highest densities observed along the Welsh Borders, Oxfordshire, the South West Peninsula and in the North. Pig density ranged from 0 to 499 animals/km2 with the highest densities observed in East Anglia and the North East.
Multivariable analysis
The results of the multivariable analysis for all sporadic cases are presented in Table 1.
Table 1. Results of multivariable analysis. All STEC O157 PTs
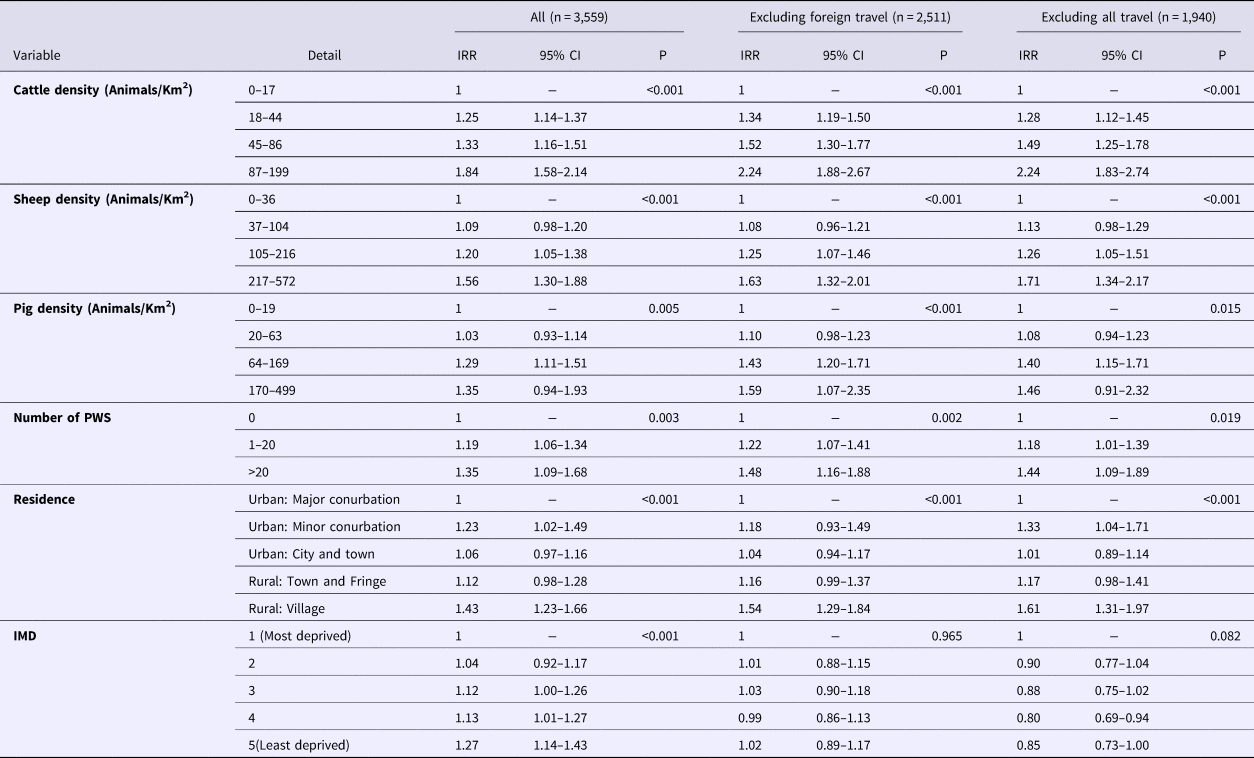
IRR, incidence rate ratio; IMD, index of multiple deprivation; CI, confidence interval; PWS, private water supplies
These indicate that living in a rural village, in an area with high densities of farmed animals (cattle, sheep or pigs), the presence of PWS and those areas considered least deprived were positively associated with risk for all sporadic cases. Removing cases reporting foreign travel removed the effect seen in the IMD variable.
The data set was then split into PT21/28 and PT8. The results for PT21/28 are presented in Table 2 and indicate that living in an area with high densities of farmed animals and being served by PWS were positively associated with risk. Living in a rural village was a risk factor only for cases who reported no travel. Areas regarded as the most deprived were associated with increased risk of PT 21/28 infection, significant only for those reporting no travel.
Table 2. Results of multivariable analysis–STEC O157 PT21/28
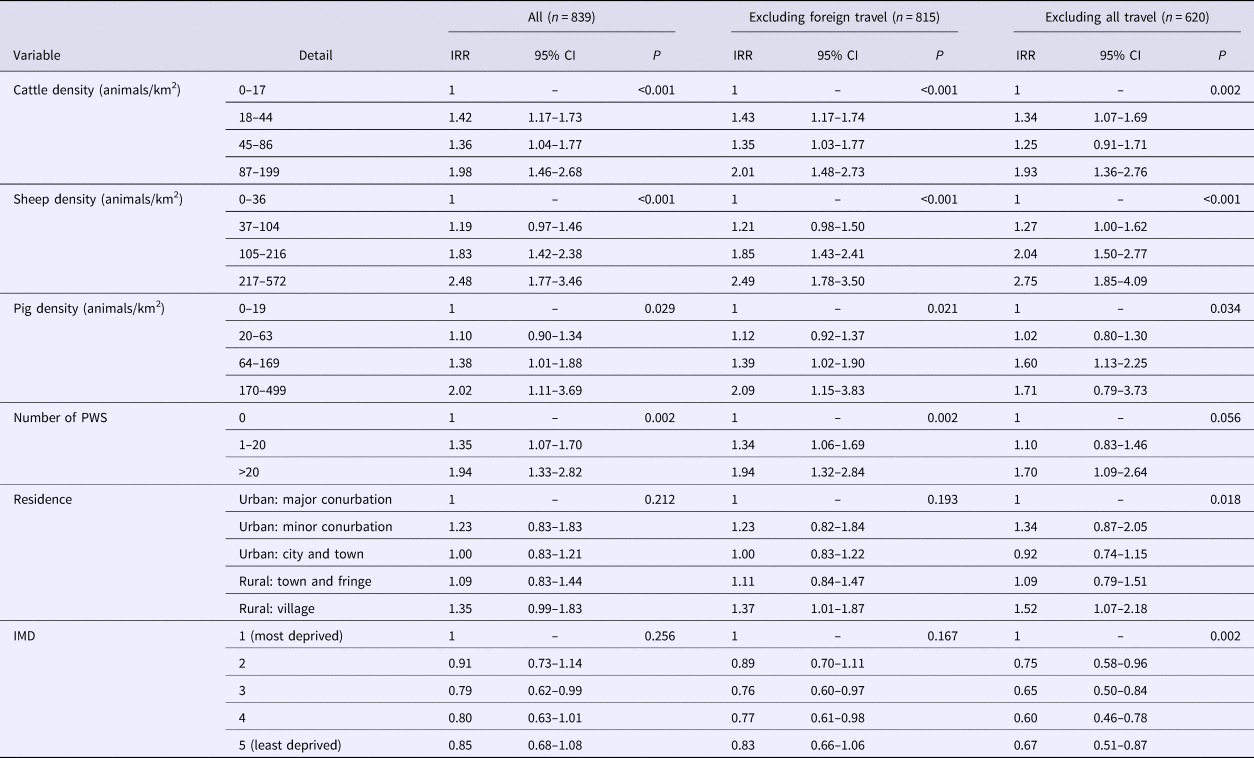
IRR, incidence rate ratio; IMD, index of multiple deprivation; CI, confidence interval
The results of the multivariable analysis for PT8 are presented in Table 3 and indicate that living in a rural village and areas with high densities of cattle and/or pigs and areas considered least deprived (quintiles 4 and 5) were positively associated with risk. Removing cases reporting foreign travel from the dependent variable increased the risk effect of cattle and pig density but removed the effect of deprivation. PWS and sheep density were not significant predictors of infection with PT8.
Table 3. Results of multivariable analysis–STEC O157 PT8
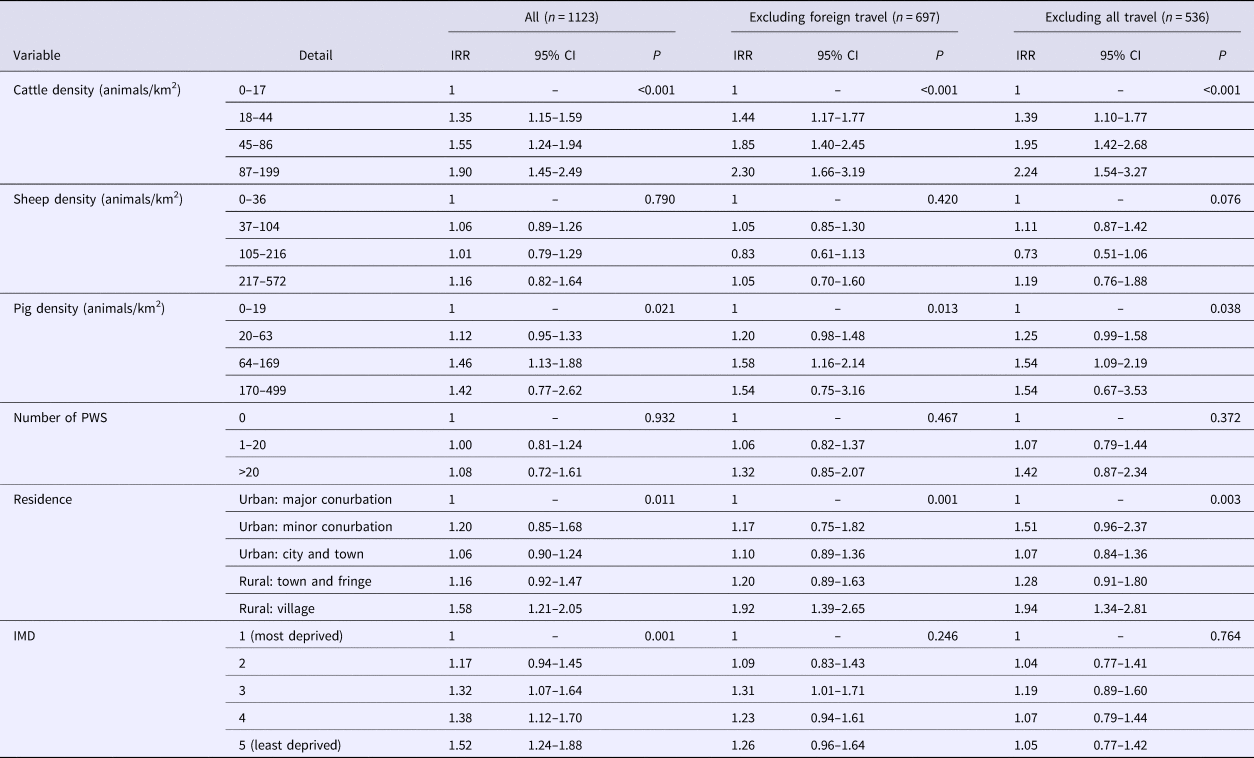
IRR, incidence rate ratio; IMD, index of multiple deprivation; CI, confidence interval
Discussion
Risk was positively associated with cattle density across all models. The risk of a case occurring in areas with 87 animals/km2 or more was more than twice that of area with fewer than 18 animals/km2. This finding was somewhat expected as cattle are regarded as the main reservoir of STEC O157 [Reference Persad and LeJeune7] and the most common subtypes shed in cattle faeces are PT21/28 followed by PT8 [Reference Milnes9].
Sheep density was positively associated with risk for all STEC O157 cases and PT21/28 cases but not for PT8. The greatest effect of sheep density was seen in the PT21/28 model where the risk was increased almost threefold in areas with high densities of sheep. There is increasing evidence that sheep and other small ruminants are an important reservoir of STEC and that sheep density is associated with non-O157 STEC infections [Reference Ohaiseadha29]. The association with PT21/28 is interesting because the carriage of PT21/28 in sheep is low (14%) compared to cattle (37%) [Reference Milnes37]; yet, they appear strongly significant in our model. This exposure to sheep and lambs has been linked to at least two PT21/28 outbreaks at petting farms/lambing events in England and an extended outbreak of closely related strains was linked to an ovine source [Reference Mikhail38, Reference Mikhail39]. A recent study in the Republic of Ireland demonstrated a geographical association between sheep density and STEC O26 but not STEC O157 [Reference Ohaiseadha29]. Disentangling the relative contribution of ruminant species to the overall burden of infection is difficult due to scant contemporary information on shedding by sheep compared to cattle, a lack of genetic difference between cattle and sheep strains [Reference Strachan40] and the fact that sheep and cattle are farmed in the same geographical areas in the United Kingdom. However, our results suggest that the role of sheep as a reservoir and potential source of infection in humans should not be overlooked.
Pig density was positively associated with risk across all models. However, for the PT8 and PT21/28 models, the observed effect was not linear. Pigs can shed STEC [Reference Cha41], and pork products have been implicated as the source or vehicle of infection in outbreaks worldwide, but they are not generally considered to be an important reservoir for STEC O157 [Reference Tseng42] or source for human infection [Reference Mughini-Gras43]. Studies of intestinal carriage in England showed a low carriage rate in pigs and that the characteristics of pig strains differed from those seen in humans during the same time period [Reference Milnes9, Reference Milnes37, Reference Chapman44]. Pig density was not identified as a risk factor for STEC infection in the Netherlands [Reference Friesema24]. In spite of this, associations with pig density appear in this study. We suggest that this finding is may be because the presence of pigs is a proxy measure for rurality and that residents in rural areas are more likely to be exposed to STEC from ruminants or other environmental sources. Compared to ruminant species, there is little evidence that pigs play an important role in the transmission of STEC O157 in England.
We found an increased risk associated with the presence of PWS for all STEC O157 cases, PT21/28 cases but not PT8. PWS that do not meet the requirements of the EC directive present a high risk of infection with STEC [Reference Risebro, Pitchers and Hunter20], however, the reason for the difference between PTs is unclear and may relate to factors not considered by this study.
Living in a rural village was a risk factor for all STEC O157 cases and for PT8. For the PT21/28 model, living outside a major urban conurbation was a significant risk factor but only for cases reporting no travel. Residents of rural areas are more likely to come into contact with contaminated environments either through work or leisure [Reference Pearl28] and residential proximity to the ruminant reservoir also increases the possibility of exposure from wildlife and insect vectors [Reference Nielsen45, Reference Nichols46].
Living in less deprived areas was strongly associated with all STEC O157 cases and PT8. What is intriguing was that when foreign travel cases were dropped, this effect was removed for all STEC O157 cases and PT8. This indicates that the deprivation effect is strongly driven by foreign travel and that risk factors for these groups differ from indigenously acquired infection. The strong association with foreign travel in the PT8 model was anticipated as a greater proportion of cases of PT8 report travel abroad compared to other PTs [Reference Byrne1]. Lower deprivation was protective for cases of PT21/28 – reporting no travel, but the reasons for this are unclear. One explanation could be related to deprivation and/or likelihood of exposure to PT21/28 compared to other strains. PT21/28 is a strain indigenous to the United Kingdom and rates of infection are highest in the north of England where there are also more areas considered to be the most deprived. However, crude rates of infection with STEC are lower in the most deprived areas, and cases are less likely to travel abroad or within the United Kingdom [Reference Adams47]. In addition, levels of social interaction differ from residents of less deprived areas [Reference Rotheram48]. Socioeconomic status has been shown to be associated with risk for other gastrointestinal infections [Reference Lake49–Reference Newman51] introducing the possibility that whilst risk factors may differ broadly across the country, within regions, socioeconomic status has a greater influence on risk factors and transmission dynamics. This is an area that would benefit from further research.
In developed countries, disadvantaged children, but not adults, appear to be at greater risk of gastrointestinal infection and that living in deprived areas is protective for infectious intestinal disease (IID) overall but is associated with more severe symptoms in those who do become infected [Reference Adams52]. Our findings indicate that once the effect of foreign travel is removed, deprivation has little effect on sporadic infection with STEC O157. This suggests that infection is a result of a localised and stochastic process driven by exposure to the local environment and that exposures related to affluence, such as diet, occupation or leisure pursuits are likely to be less important.
Residential distance from the coast and living in an area with a high proportion of fresh water coverage were not significant and removed at an early stage of the modelling process. These environments may act as reservoirs for STEC [Reference Williams53] and recreational exposure to fresh water has been suggested as a risk factor for STEC infection in epidemiological studies from other countries. The lack of an association in the United Kingdom may relate to the general unpopularity of freshwater swimming in the United Kingdom in comparison to other countries [Reference Joosten54–Reference Marion57]. These variables were proxies for exposure and so do not capture details of individual interactions with these environments.
There are several potential limitations to our study. First, molecular typing methods, used routinely in England since 2015, are superior to the phenotypic methods we used to discriminate between cases and have been shown to reduce the number of cases considered to be sporadic [Reference Byrne58]. It is therefore possible that a small number of cases included in our study were microbiologically linked and therefore may not be considered truly sporadic. Second, we excluded cases linked to household outbreaks. We made this decision based on the difficulty in identifying the primary case amongst co-primary and asymptomatic (microbiologically confirmed) cases generated by microbiological screening of household contacts. In addition, we noted epidemiological links between foodborne outbreaks and secondary transmission within households which may have introduced a bias away from the exposures of interest. Third, for every STEC O157 infection reported to national surveillance systems in England, there are an estimated 7.4 in the community [Reference Tam59]. The reasons for this are likely to be related to severity of disease, health-seeking behaviours and whether or not a clinician takes a sample and requests a microbiological examination from a laboratory. Notwithstanding that the ratio of STEC O157 reports is considerably smaller than other pathogens; the cases reported to national surveillance represent a biased sample of the true community burden of STEC O157 in England. Transmission routes are varied and infection is a result of complex interactions between people and their local environment. Our approach meant that individually reported exposures could not be considered in our analysis. Finally, this is an ecological study and association does not equal causation which could only be inferred from other study designs involving an intervention. We are confident that the association with animal density is most likely due to environmental exposure, however, other factors not included in our study (e.g. locally sourced food) cannot be ruled out as a potential route of infection.
In conclusion, using arguably one of most comprehensive enhanced surveillance of STEC data sets in the world, we found that two-thirds of infections were sporadic and that the spatial and temporal distribution of these cases showed distinct variation within England. We provide evidence that living in a rural area with high densities of farmed animals and served by PWS partly explain this variation. Our results indicate that travel abroad may expose individuals to risks not present in their local residential environment and that this risk is influenced by socioeconomic status. Further analysis is required to elucidate the relative importance of exposures reported by individual cases including travel, contact with animals and the agricultural environment and consumption of food and water.
To reduce the overall burden of infection in England, interventions designed to reduce the number of sporadic infections with STEC O157 should focus on the residents of rural areas with high densities of livestock and the effective management of non-municipal water supplies.
Acknowledgements
The authors thank Marie Anne Chattaway, Vivienne DoNascimento and Neil Perry for their microbiology expertise. They also acknowledge the roles of Amy Mikhail, Lisa Byrne, Kirsten Glen and Nalini Purohit for the maintenance of the enhanced surveillance system for STEC in England and extend thanks to Sue Pennison of the Drinking Water Inspectorate for sharing their data on PWS locations. Finally, they are grateful to the microbiologists and health protection and environmental health specialists who have contributed data and reports to national surveillance systems and the epidemiologists and information officers who have worked on the national surveillance of intestinal infectious diseases for the National Infections Service of Public Health England.
Financial support
The research was funded by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at University of Liverpool in partnership with Public Health England (PHE), in collaboration with University of East Anglia, University of Oxford and the Institute of Food Research.










