Background
Shigellosis, also known as bacillary dysentery, is caused by four species: Shigella flexneri (S. flexneri), Shigella sonnei (S. sonnei), Shigella dysenteriae (S. dysenteriae) and Shigella boydii (S. boydii). Symptoms include profuse, often bloody, diarrhoea, abdominal pain and fever, with an average duration of 4–5 days. The median incubation period is 2 days with a range of 12 h to 4 days [Reference Chai1]. The infectious period is primarily during diarrhoeal illness; however, cases can remain infectious for as long as the organism is excreted in the stool (~14–28 days) [Reference Chai1].
In the UK, all four species of Shigella are a known cause of travellers' diarrhoea. S. dysenteriae and S. boydii are almost exclusively acquired outside the UK [Reference Bardsley2] but S. flexneri and S. sonnei may be indigenously acquired. Shigella species are anthroponotic (pathogens that are generally specific to human hosts) and the infectious dose is low (10–100 organisms) so person-to-person transmission is common [Reference DuPont3]. Outbreaks tend to occur within households, nursery schools, and community settings [Reference Chattaway4, Reference Dallman5]. However, the recent epidemiology has seen increased transmission amongst adult males relative to all other demographic groups and outbreaks of S. sonnei and S. flexneri have been linked to person-to-person spread amongst men who have sex with men (MSM) [Reference Bardsley2, Reference Simms6]. Food and waterborne outbreaks are rarely detected in the UK but do occur. Infected food-handlers and fresh, leafy green produce, such as salad and herbs, are the most common sources and vehicles of transmission [Reference Long7–Reference Nygren9]. The last foodborne outbreak in England was associated with iceberg lettuce [Reference Frost10].
This report describes the steps taken by the outbreak control team (OCT) to determine wider epidemiological links between cases linked to locally identified outbreaks and those that appeared sporadic but clustered by whole-genome sequencing (WGS), within the WGS cluster and to identify any common exposures.
In April 2018, local Health Protection Teams (HPTs) in the West Midlands and West Yorkshire regions of the UK independently identified two outbreaks of diarrhoea and vomiting linked to three restaurants that were geographically distinct. Microbiological investigations identified S. sonnei as the aetiological agent. Routine assessment of S. sonnei WGS clusters by the national Gastrointestinal Infections team identified further sporadic cases belonging to the same genetically related cluster but linked to different food outlets.
Methods
Microbiological investigations
The UK Standards for Microbiology Investigation of Faecal Specimens for Enteric Pathogens recommends testing of all faecal specimens for Shigella species in individuals reporting symptoms of gastrointestinal disease (https://www.gov.uk/government/publications/smi-b-30-investigation-of-faecal-specimens-for-enteric-pathogens). Approximately two-thirds of Shigella spp. isolates are submitted to the Gastrointestinal Bacterial Reference Unit (GBRU) at the Public Health England for confirmation of species identification and typing, in concordance with local practice. Many laboratories refer all isolates, whereas others may submit to GBRU only when local identification is uncertain and/or typing is required for further clinical or public health investigation. Since 2015, species identification, serotyping and molecular typing have been performed using WGS [Reference Dallman5, Reference Chattaway11].
DNA from isolates of S. sonnei submitted to GBRU was extracted for sequencing on the Illumina HiSeq 2500 instrument. High-quality Illumina reads were mapped to the S. sonnei reference genome Ss46 (Genbank accession: NC_007384.1) [Reference Wei12] using BWA MEM v0.7.12 [Reference Li and Durbin13] and Samtools v1.1 [Reference Li14] Single Nucleotide Polymorphisms (SNPs) were identified using GATK v2.6.5 [Reference McKenna15] in unified genotyper mode. Core genome positions that had a high-quality SNP (>90% consensus, minimum depth 10×, GQ > = 30) in at least one isolate were extracted and RaxML v8.2.8 [Reference Stamatakis16] was used to derive the maximum likelihood phylogeny of the isolates after first removing regions of the genome predicted to have undergone horizontal exchange using Gubbins v2.0 [Reference Croucher17, Reference Dallman18]. Genome-derived serotyping and resistance determinant detection was performed using the GeneFinder tool [Reference Dallman5, Reference Sadouki19].
Data availability statement
Sequence data is publicly available on NCBI BioProject PRJNA315192.
Epidemiological investigations
On 3 May 2018, an OCT was formed to coordinate the investigation with the aims of identifying all cases linked to the outbreak, establishing epidemiological links between the cases linked to the restaurants and those belonging to the genetically related cluster and identifying and implementing control measures to prevent further cases.
Case definitions
Cases were defined as follows:
-
Confirmed case: A laboratory-confirmed S. sonnei infection reported after 1 March 2018 belonging to clonal complex 152 5-SNP cluster 1.1.29.49.547.1303.%
-
Probable case: A resident of West Yorkshire or the West Midlands with a laboratory-confirmed S. sonnei infection with an onset date after 1 March 2018 and an epidemiological link to an implicated venue.
-
Possible case: A resident of West Yorkshire or the West Midlands with symptoms of diarrhoea and/or vomiting and an onset date after 1 March 2018 and with an epidemiological link to an implicated venue or a confirmed/probable case.
-
Secondary case: A member of the same household as a confirmed, probable or possible case with no exposure or link to the implicated venues who became symptomatic and/or tested positive for S. sonnei.
Retrospective cohort study
The national OCT recommended that a retrospective cohort study be conducted to test the hypothesis that illness was associated with consumption of one or more food items or ingredients served at Venue 3, where the majority of cases reported eating, on 29 March 2018. For the purposes of this study, a case was defined as a guest or staff member who attended or worked at the associated restaurant (Venue 3) on 29 March 2018 and who developed symptoms of diarrhoea and/or abdominal pain with fever within 4 days after eating at Venue 3, without an alternative explanation, or had Shigella spp. detected in a faecal specimen.
Data were collected through a standardised online questionnaire. Questions included demographic information (age, sex and occupation), information on symptoms and symptom onset and duration, the severity of the disease, history of illness prior to the event, any possible secondary transmission, travel history, food and water consumed at the event, risk exposures related and other events attended that could explain illness amongst the guests.
Environmental Health Officers (EHOs) collected information on food items served at Venue 3 on 29 March 2018 including the details of each ingredient used per dish. No complete guest list with all 22 attendees was available, and guests with contact information were asked to forward the questionnaire to other attendees, regardless of whether or not they were ill. The questionnaire was piloted by FS West Midlands colleagues and other members of the OCT. The questionnaire was open for 21 days due to a low response rate. A reminder was sent after 7 and 14 days, respectively. All data were collected, cleaned and analysed using Microsoft Office Excel and Stata version 14.0 (Stata Corp., College Station, TX, USA).
Food chain investigations
Forward and backward food chain information was collected for all ingredients of meals eaten by cases at the four venues linked to two or more confirmed cases with exposure dates in March 2018. Data collection and management were performed using FoodChain-Lab [Reference Weiser20] and tracing scores were calculated for venues and products under investigation to identify common supply chains as previously described [Reference Weiser20].
Microbiology of food and environmental specimens
All food specimens collected as part of this outbreak investigation were examined with the British Standards Institution (2004) BS EN ISO 21567:2004 – Microbiology of food and animal feeding stuffs – horizontal method for the detection of Shigella species. Testing was conducted at the PHE Food, Water and Environment (FW&E) laboratory in Colindale, London.
Results
Descriptive epidemiology
A total of 33 cases, linked to 7 different venues were identified during the investigation (Table 1). Of these, 14 met the confirmed case definition of which 13 were primary and one was the secondary case. A further 19 met the possible case definition. Enhanced surveillance questionnaires were completed for all confirmed cases.
Table 1. Number of cases associated with each linked venue

The majority (64%) of confirmed cases were male and the mean age was 34 years (range: 12–59 years) (Fig. 1). All confirmed cases reported symptoms of diarrhoea followed by fever (92%), vomiting (75%) and bloody/mucoidal stools (42%). Possible cases reported acute onset vomiting and diarrhoea within 2 days of exposure to Venue 3 (n = 11) or Venue 4 (n = 4). Symptom onset dates for the bulk of cases ranged from 26 March to 3 April 2018 (Fig. 2). Four cases were hospitalised for between two and five nights.
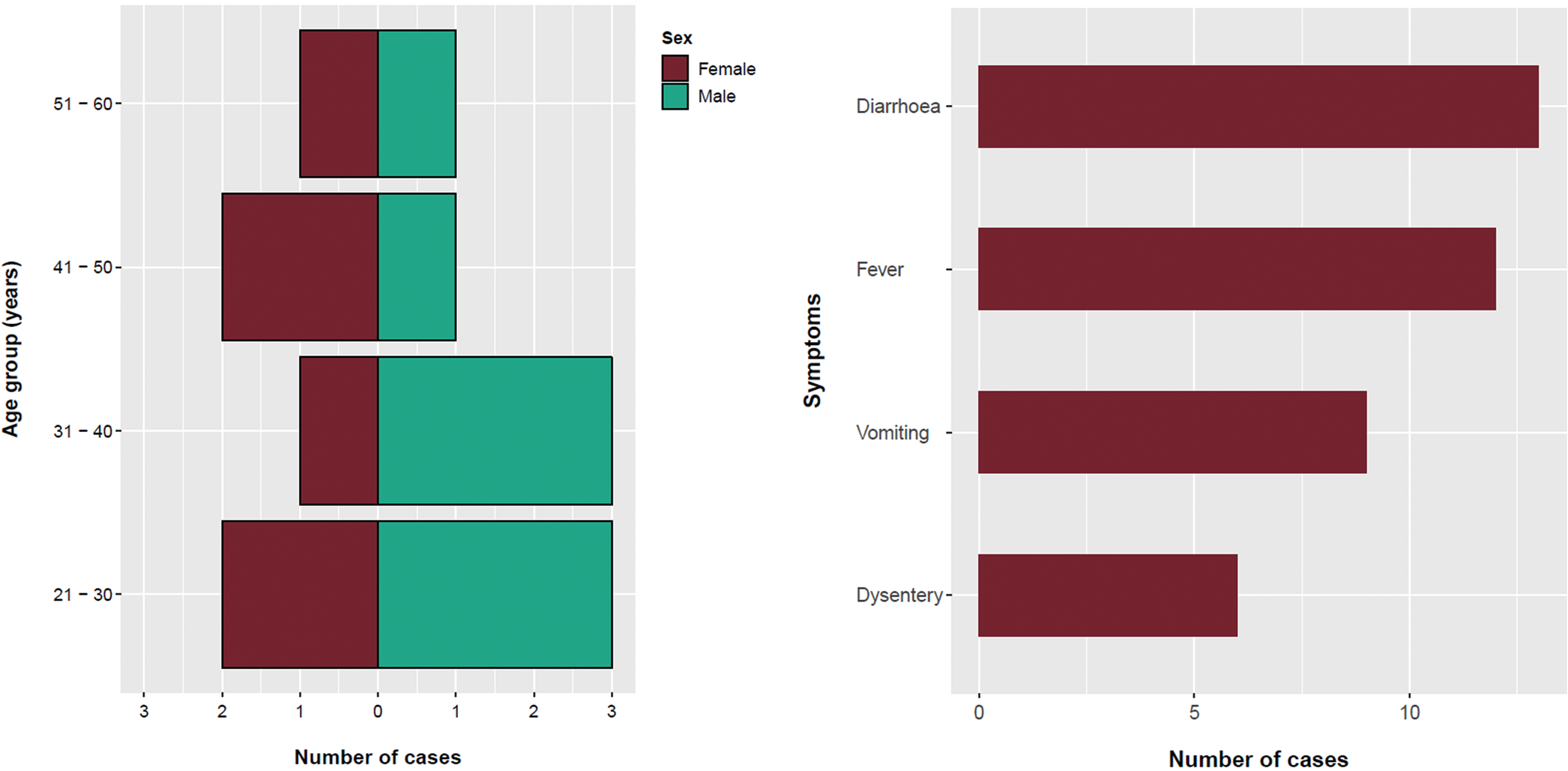
Fig. 1. Age, sex and symptom distribution of cases, England, March–May 2018.
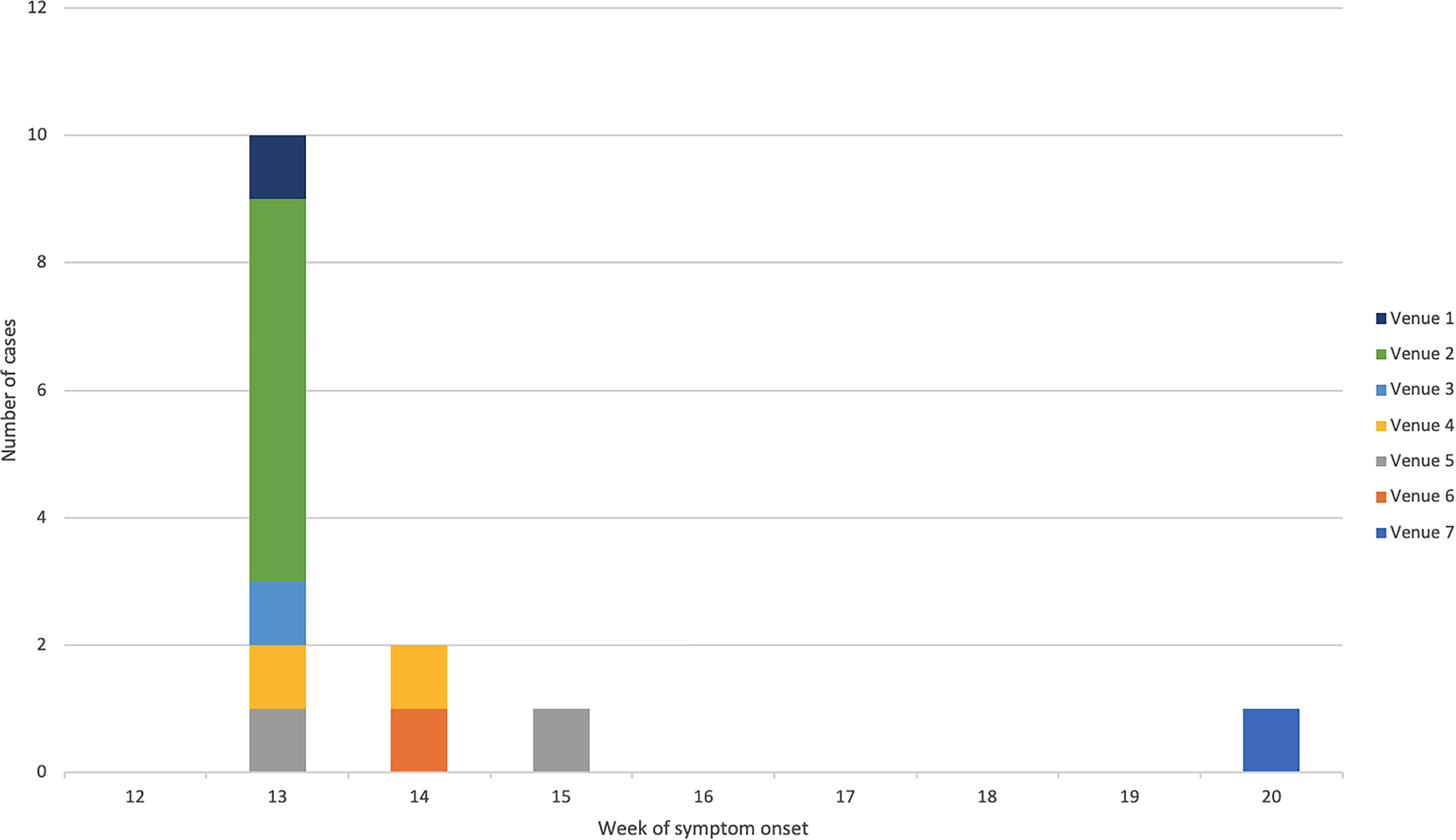
Fig. 2. Epidemic curve of confirmed cases by week of symptom onset and linked venue.
Cases were exposed at the implicated venues between 24 and 31 March 2018. The earliest exposure occurred at Venue 1 in Bedford, followed by the venues in the West Midlands Venues 2 and 3 and the latest exposures were in Bradford Venues 4 and 5. The median time between exposure and onset was 2 days (range: 0–4 days). A further case, identified in May 2018, reported eating fresh coriander at home and shopping at a supermarket specialising in Middle Eastern food products. This supermarket also had a food outlet serving shawarma/kebab type dishes and also supplied the local catering trade with ingredients. Most cases lived close to the outlets where they were most likely infected. There was no evidence of any movement of food handlers between implicated venues.
Microbiological investigations including WGS
Of the 17 cases that submitted stool specimens, isolates from 14 were submitted to GBRU for laboratory confirmation and WGS. All 14 belonged to the same 5-SNP single linkage outbreak cluster that fell within a 25-SNP cluster comprising 177 isolates submitted to GBRU between June 2015 and May 2018. Of those in the wider cluster, 63/177 (35.6%) reported travel abroad within seven days of onset of symptoms, most of whom 51/63 (81.0%) reported travel to Pakistan (Fig. 3). Laboratory recording of foreign travel is incomplete, so the proportion of cases that acquired their infection outside the UK may be higher.

Fig. 3. Phylogenetic tree of S. sonnei at the 25-SNP single linkage cluster. The blue insert highlights the 5-SNP single linkage cluster, including historical cases from 2017. The pink insert highlights cases from 2018 meeting the confirmed case definition. Annotations include the short-read accession number (SRR number), travel destination if stated, and presence of CTX-M-15.
Prior to the outbreak, this clade was phylogenetically heterogeneous; in contrast, all case isolates in the outbreak investigated here fell within a 5-SNP single linkage cluster designated 1.1.29.49.547.1303.% and the majority 10/14 (71%) were 0 SNPs apart. Only one confirmed case in the outbreak cluster reported recent travel, returning to the UK 3 days prior to becoming ill (Fig. 3). A review of earlier cases on the WGS database identified five cases from 2017 that fell within the same 5-SNP cluster: all had links to Pakistan.
Antimicrobial resistance profiles extracted from WGS data indicate that the outbreak strain had the following resistance genes or mutations: extended-spectrum beta-lactamase (ESBL) bla CTX−M−15, a single mutation in gyrA D87Y, and a plasmid-encoded fluroquinolone resistance gene qnrS1, streptomycin (strA, strB), trimethoprim (dfrA1), tetracycline (tetA) and sulphonamide (sul2). Phenotypic antimicrobial sensitivity testing confirmed that this strain was an ESBL-producer, susceptible to azithromycin and intermediate or resistant to ciprofloxacin (MIC 0.25−5.0 mg/l), and was resistant to streptomycin, trimethoprim, tetracycline and sulphonamide. One hospitalised case had evidence of treatment failure with ciprofloxacin and was subsequently treated with meropenem.
Food trace-back investigations
Food chain trace-back information was collected for ingredients of dishes served to cases from three of the four affected premises. Limited information was available for ingredients supplied to Venue 1, and not all food items from the menu were included in the trace-back investigation.
Four ingredients (chicken, green chillies, fresh coriander and onions) were served at all four restaurants selected for the FoodChain-Lab investigation. No single common supplier or ingredient was identified. Venues 2, 3 and 4 reported purchasing fresh coriander from local markets and Venue 1 reported that they bought theirs from a national supermarket chain. It was not possible to identify where the coriander was grown.
All venues linked to this investigation served kebab/shawarma/gyros with salad items (tomatoes, cucumber, onions, iceberg lettuce, fresh chillies, jalapeno peppers), pickles and sauces (chilli sauce, tahini sauce, mint yoghurt, mayonnaise). Limes and fresh coriander leaves were also added to salads or used as a garnish. Venue 1 served Thai food and although coriander was not listed as an ingredient on their menu, photographs were available on used coriander as a garnish.
Cohort study results
Of 22 guests attending Venue 3, 8 completed the questionnaire, representing a response rate of 36.4%. Of the eight respondents, seven (87.5%) were symptomatic and eligible for inclusion in the study but were not laboratory-confirmed cases. However, only one person who was not ill responded so the following results are descriptive only.
The epidemic curve indicated a point-source exposure with illness peaking between 1 and 2 days after exposure. Based on the timing of food consumption among guests (i.e. 16:30 h), the median incubation period was 32.5 h (mean: 35.6 h, range: 17.5–67.5 h).
Of the eight respondents, 62.5% were females (n = 5), and the median age of the respondents was 47.0 years (mean: 44.3 years; range: 21–56 years). All cases reported fatigue, 85.7% reported abdominal pain, or nausea, 71.4% reported diarrhoea, fever, muscle ache or headache.
The majority of cases reported eating the sheesh kebab (85.7%) and/or chicken tikka wraps (85.7%). The sheesh kebab was the only dish that contained fresh coriander. Due to poor participation, the statistical inference could not be drawn from the study.
Environmental investigation
EHOs noted poor temperature control, poor cleaning standards, lack of hand hygiene facilities at Venue 3 which had a food hygiene rating of 1 within a range of 0 (worst) to 5 (best) indicating that major improvement was necessary (https://www.food.gov.uk/safety-hygiene/food-hygiene-rating-scheme). No staff members were reported to have been unwell. No food samples were taken because perishable food items served on 29 March were already consumed or had been discarded prior to the inspection. Following the outbreak, the restaurant was refurbished, and advice was given on kitchen routing improvements. Food Standards Agency advice on good food preparation practices was also provided to staff https://www.food.gov.uk/business-guidance/personal-hygiene.
Discussion
The national foodborne outbreak of S. sonnei described here was linked to food exposures at multiple restaurants located in different geographical areas that were not part of a franchise; WGS facilitated the identification of potential links between these restaurants. Analysis of WGS data also demonstrated a close phylogeographical association between the outbreak strain and isolates from UK cases reporting recent travel to Pakistan.
Food traceback investigations revealed that fresh coriander leaves were the only common ingredient supplied to all venues attended by cases.
Foodborne S. sonnei outbreaks are uncommon in the UK. The majority of outbreaks described in the literature have been associated with contaminated fresh produce, including salad vegetables and herbs, which are known to support the growth and maintenance of Shigella spp., especially at refrigeration temperatures [Reference Long7–Reference Nygren9, Reference Waldram21]. These include an EU-wide outbreak due to iceberg lettuce contaminated with human sewage during harvest in Spain [Reference Frost10, Reference Kapperud22] and baby sweet corn contaminated in a packing shed in Thailand that resulted in cases in Denmark and Australia [Reference Lewis23]. Imported fresh herbs such as parsley, basil and coriander have also been implicated in multiple S. sonnei outbreaks, typically in association with restaurant meals where uncooked herbs were used as a garnish [Reference Guzman-Herrador8, 24]. Fresh coriander leaves from South East Asia were implicated in an S. sonnei outbreak in Sweden in 2015 (https://www.foodnavigator.com/Article/2015/11/25/Sweden-finds-coriander-to-be-source-of-shigellosis). This outbreak had several features in common with the outbreak described here, specifically that cases had attended multiple restaurants in two regions in Sweden and were only linked following WGS of their isolates. Fresh coriander leaves have been implicated in S. sonnei outbreaks in the USA (http://www.outbreakdatabase.com/details/senor-felix-5-layer-bean-dip-2000/?organism=Shigella). Contamination of fresh produce with Shigella spp. in South East Asia has also been highlighted; a study of gastrointestinal bacterial contamination in Indian street food found that 6% of coriander sauces tested were contaminated with Shigella spp. [Reference Ghosh25].
We considered three scenarios that could explain the role of fresh coriander as the vehicle of infection for this outbreak These were (i) the coriander was contaminated at the point of production, (ii) contamination occurred during the wholesale distribution of the coriander and (iii) infected food handlers contaminated the coriander in restaurants. The outbreak control team concluded that the first and second scenarios provided the most plausible explanations for this outbreak. Bulk supplies of coriander entering the wholesale market (irrespective of source) are broken down into smaller batches or bunches at multiple locations. This is performed by hand, providing an opportunity for contamination by an infected food handler. Although subject to general food law, fruit and vegetable wholesalers are not generally considered to present a high risk in terms of food safety. As such, their food safety systems and procedures are unlikely to be as sophisticated as those of higher-risk businesses. The third explanation is less plausible as there was no evidence that food handlers had links with more than one food outlet and none had reported symptoms of gastrointestinal infection. We were unable to confirm whether any of the food handlers had recently travelled outside the UK.
Because of the time-lag between the local identification of outbreaks, confirmation by WGS and the identification of coriander leaves as a potential vehicle of infection, coriander leaves were not sampled for testing as part of the initial outbreak investigations. Subsequently, a decision was taken not to sample coriander as part of the national investigation as too much time had lapsed following case exposure, no new cases were detected during the investigation and the implicated product was likely no longer in circulation due to its short shelf life. Shigella spp. were not isolated from other fresh produce sampled as part of local investigations. However, the microbiological testing is culture-based and not molecular testing, which may lack the sensitivity to detect low-level contamination in food. In addition, food samples were collected 6 and 9 days after case exposures, respectively, rendering it unlikely that the products tested were the same batch as the products that cases had consumed prior to the onset of their illness.
AMR data derived from the WGS profile rapidly determined the MDR profile of the outbreak strain, highlighting the risk of potential treatment failures caused by the reduced susceptibility to ciprofloxacin which is a common first-line agent used in the UK for severe shigellosis. Although third-generation cephalosporins are not recommended as the first-line drugs for the treatment of shigellosis in the UK, the presence of bla CTX−M−15 encoded on a mobile genetic element is associated with a risk of carriage and transmission of resistance, to other bacteria within the host gastrointestinal tract. Complicated and immunocompromised patients are likely to have higher morbidity as demonstrated by at least one case who needed to be treated with a carbapenem. S. sonnei harbouring bla CTX−M27 has been associated with outbreaks of GI symptoms in MSM [Reference Mook26] but has not been previously described in foodborne outbreaks in the UK.
This outbreak highlights the potential for a multidrug-resistant strain of S. sonnei to be transmitted via a food vehicle that is distributed over a wide geographic area. Fresh produce, such as salad items and fresh herbs, have a short shelf life and the contaminated batch is often not available for microbiological testing by the time a likely vehicle has been identified. Although food traceback is challenging for identifying stealth vehicles, defined as minor components of a meal which cases may not recall having eaten, as a potential cause of foodborne illness, it is essential for outbreak investigations [Reference Byrne27]. Routine, prospective collection of enhanced surveillance questionnaires of all S. sonnei cases in England could help quantify the role of food as a risk factor for shigellosis in England. Combined with WGS, this information would facilitate the rapid detection of foodborne outbreaks, enhance traceback investigations and increase the probability that vehicles of infection are correctly identified.
Acknowledgements
The authors would like to thank the members of the national and local outbreak control teams for their contributions. They are (in alphabetical order): Neil Anstey, Nachi Arunachalam, Gavin Bailey, Srilaxmi Degal, Roger Gajraj, Janine Gos, Gareth Hughes, Eunice Kwok, Katherine Martin, Jim McLauchlin, Richard Puleston, Yasmin Rehman, Adam Spencer, Mamoona Tahir and Rehman Teagle
Financial support
RE, JH, DRG and CJ are affiliated to the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at the University of Liverpool in partnership with Public Health England (PHE), in collaboration with the University of Warwick, and are based at PHE. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health and Social Care or Public Health England.
Conflict of interest
There are no conflicts of interest.







