INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) are important human pathogens that can cause a variety of clinical manifestations, from self-limited diarrhoea to more severe illness such as haemorrhagic colitis or haemolytic uremic syndrome (HUS) [Reference Karmali, Gannon and Sargeant1]. Hence, STEC is the most common infectious agent causing HUS, a syndrome characterised by progressive renal failure. HUS is associated to high morbidity and mortality during the acute phase and to long-term renal and extrarenal complications, particularly in children [Reference Scheiring, Andreoli and Zimmerhackl2]. Ruminants, and cattle in particular, are considered the primary reservoir of STEC, and the main source of human infection [Reference Mughini-Gras3]. Contamination of food of animal origin can occur during milking, slaughtering and evisceration. Moreover, faecal shedding of STEC by infected animals can contaminate the environment of the farm, where STEC may survive for long periods of time [Reference Maule4]. Contaminated manure used as fertiliser or effluents from farmland containing STEC can also contaminate fresh produce [Reference Solomon, Yaron and Matthews5].
STEC are distinguished from non-pathogenic E. coli strains isolated from the normal intestinal microbiota of healthy mammals by the production of one or more variants of Shiga toxin (Stx), encoded by stx1 and/or stx2 genes. The presence of stx2 has more frequently been associated with severe disease [Reference Paton and Paton6]. Aside from the production of Stx, STEC have other potential virulence factors such as the intimin, an outer membrane protein encoded by the eae gene that facilitates intimate attachment to intestinal epithelial cells thereby increasing disease severity [Reference Donnenberg7], and the plasmid-encoded enterohaemorrhagic E. coli haemolysin (EhxA)[Reference Lorenz8]. Several O serogroups have been found associated with Stx production, but O157:H7 is considered the most pathogenic serotype and is responsible for most HUS cases of bacterial aetiology worldwide [Reference Mainil and Daube9]. In addition, other non-O157 STEC serogroups have also been implicated in severe disease forms as, for example, O111, a serogroup associated with HUS and frequently incriminated in outbreaks in Spain [Reference Mora10] and worldwide [Reference Piercefield11].
In Spain, declaration of human STEC isolations has only recently become compulsory (Order SSI/445/2015), as per European Union legislation, making the reporting of human STECs to European Food Safety Agency (EFSA) only partial and based on sentinel surveillance so that no information on estimated coverage and notification rate can be estimated. Furthermore, the monitoring programmes of STEC in ruminants are performed only in young beef cattle (1–2 years old) at slaughterhouses selected from different regions of Spain to represent the total volume of cattle sacrificed in the country. As such, small regions like the Basque Country (northern Spain) are not well represented and so the real prevalence in livestock of STEC in the Basque Country cannot be inferred from those surveys. Furthermore, the last study performed regarding livestock incidence of STEC in the Basque Country was carried out in 2003–2005 and in that study STEC was shown to be widespread in cattle and sheep [Reference Oporto12]. Moreover, notification compliance rate of human STEC cases in the Basque Country has been traditionally higher than in other regions and comparable data are available in annual reports. Based on those reports, the incidence of STEC infection in humans has been increasing over the years (0.28 in 2004 [13], 0.51 in 2015 [14]) culminating in an incidence of 1.11 cases/100 000 inhabitants in 2016 [15]. The absence of recent studies regarding the prevalence of STEC in livestock precludes the inference of the contribution of livestock in the increase of STEC human cases in our region.
The use of antibiotics in the treatment of STEC infections is controversial and not recommended according to the current clinical guidelines, as certain antimicrobials can induce Shiga toxin production [Reference Wong16, Reference Kimmitt, Harwood and Barer17]. However, antimicrobial resistance (AMR) is a subject of growing concern due to the widespread of E. coli resistant to all antibiotics used in human therapy, and the dissemination of resistance through mobile genetic elements. A recent example is the detection in China of plasmid-mediated antibiotic resistance to colistin [Reference Liu18], one of the last resource antibiotics for the treatment of multi-drug-resistant Enterobacteriaceae in humans, which has recently been also isolated in cattle in Spain [Reference Hernández19]. Thus, systematic monitoring of the occurrence of AMR in both commensal and pathogenic E. coli, like STEC, is essential to measure the spread of resistance among food-borne bacteria and design and implement new control strategies [Reference Threlfall20].
In this context, the objectives of the present study were (i) to update herd-level prevalence estimates of STEC in their main animal reservoirs (cattle and sheep) in the Basque Country; and (ii) to assess the potential health risk for humans of livestock-carrying STEC through the analysis and comparison of virulence and AMR profiles of human and ruminants STEC strains.
METHODS
Sampling design for the estimation of STEC prevalence in ruminant farms
A cross-sectional survey was carried out to estimate the prevalence of STEC in cattle herds and sheep flocks in the Basque Country (Northern Spain). The sampling strategy has been previously described [Reference Hurtado, Ocejo and Oporto21], as this study is part of a larger one that intends to determine the prevalence of Salmonella, Listeria monocytogenes [Reference Hurtado, Ocejo and Oporto21] and thermophilic campylobacters (unpublished data) in ruminant farms. Briefly, the number of herds of beef cattle, dairy cattle and sheep farms to be sampled was determined based on the official census for an expected herd prevalence of 50%, a 95% confidence level and an accuracy of 10% using Win Episcope 2.0. Thus, a total of 301 herds (115 dairy sheep, 104 beef and 82 dairy cattle) were sampled once between February 2014 and June 2016. Data on herd size were collected. Rectal faecal samples from 25 animals randomly selected per herd were collected with a gloved hand, and a 25 g pool was prepared (1 g per animal per pool) for microbiological analyses. Sample collection was carried out by veterinary practitioners as part of the usual screening scheme performed on farms, strictly following Spanish ethical guidelines and animal welfare regulations (Real Decreto 53/2013). The collection of this material, being considered routine veterinary practice, did not require the approval of the Ethics Committee for Animal Experimentation. Informed oral consent was obtained from the farm owners at the time of sample collection. Details on general husbandry systems for beef cattle, dairy cattle and sheep in the Basque Country were reported elsewhere [Reference Hurtado, Ocejo and Oporto21].
STEC isolation from animal faeces and strain characterisation
Faeces (25 g of pooled faecal samples) were diluted 1:10 in modified Tryptic Soy Broth (mTSB, BioMérieux) supplemented with novobiocine (Biolife) and incubated at 41 ± 1 °C for 6–7 h. The mTSB broth was subcultured onto MacConkey agar at 37 ± 1 °C for 24 h. To detect STEC, 10 lactose-positive colonies from the MacConkey agar were analysed in two pools of five in a multiplex Real-Time PCR reaction targeting the genes encoding the Shiga toxin 1 and 2 (stx1 and stx2) and the E. coli attaching and effacing gene (eae). For stx-positive pools, colonies were individually tested to confirm their identity as STEC using the same multiplex Real-Time PCR. Primers and probe for eae were as previously described [Reference Nielsen and Andersen22], whereas those for stx1 and stx2 were based on those described by Perelle et al. [Reference Perelle23] with slight modifications in the sequence of the primers (stx1-F: 5′-TTTGTTACTGTGACAGCTGAAGCTTTA-3′; stx1-R: 5′-CCAGTTCAATGTAAGATCAACATCTTC-3′; stx2-F: 3′-GTCACTGTCACAGCAGAAGCCTTA-5′; stx2-R: 5′-CAGTTCAGAGTGAGGTCCACGTC-3′). One STEC isolate from each positive herd/flock was selected for further analyses, with the exception of herds/flocks where more than one stx1/stx2/eae profile was found. Further characterisation included the amplification of the genes encoding the O157 somatic antigen (rfbE), the H7 flagellar antigen (fliC) and the enterohaemolysin (ehxA) in a conventional multiplex PCR [Reference Wang, Clark and Rodgers24], and the O111 serogroup-related gene wzxO111 [Reference Sánchez25].
The detection of O157 STEC was performed as follows: DNA was extracted directly from the bacterial culture obtained from MacConkey agar plates and screened for the presence of the rfbE gene by Real-Time PCR as previously described [Reference Perelle23]. PCR-positive samples were then submitted to immunomagnetic separation (IMS) using magnetic Dynabeads anti-E. coli O157 (Dynal, Oslo, Norway), followed by chromogenic isolation (chromID™ O157:H7/NM agar with CT, BioMérieux). Typical bluish-green colonies (two per sample) were considered presumptive E. coli O157 and confirmed by Real-Time PCR amplification of rfbE as described above. Confirmed E. coli O157 colonies were characterised for the presence of stx1/stx2/eae (multiplex Real Time PCR) and fliC and ehxA (conventional PCR) using the abovementioned procedures for STEC.
Minimum inhibitory concentrations (MIC) were determined by broth microdilution using a Sensititre® MIC Susceptibility Plates (EUVSEC1, ThermoFisher Scientific, Waltham, MA, USA) containing 14 antimicrobial agents (12 classes) following recommendations by the Commission Decision 2013/652/EU. Results were interpreted using epidemiological cut-off values based on the distribution of MICs of wild-type susceptible populations as developed by the European Committee for Antimicrobial Susceptibility Testing (EUCAST, http://www.eucast.org).
STEC isolation from human stool samples and strain characterisation
A total of 29 094 stool samples from inpatients and outpatients of all ages sent to the Microbiology Service of Hospital Universitario Donostia between January 2014 and December 2016 with symptoms of gastrointestinal disease were screened for the presence of STEC. Following routine procedures, DNA was automatically extracted from stool samples (NUCLISENS® EASYMAG®, BioMérieux) and analysed using a real-time multiplex PCR kit (FTD Bacterial gastroenteritis, Fast Track diagnostic®, Werfen) that detects Salmonella spp., Shigella spp., Yersinia enterocolitica, Clostridium difficile, Campylobacter (C. coli/C. jejuni/C. lari) and STEC (stx+). Stx-positive stool samples were cultured onto SMAC and MacConkey agar (BioMérieux) and incubated at 37 ± 2 °C for 24–48 h. Typical sorbitol-negative E. coli O157 colonies on SMAC agar were identified by MALDI-TOF and confirmed by O157-specific slide agglutination using the E. coli O157 Latex Test Kit (Oxoid), and further characterised by PCR for the presence of stx1, stx2, eae, ehxA and rfb O157 as previously described [Reference Paton and Paton26]. One lactose-positive colony per MacConkey agar plate was also analysed by MALDI-TOF, and those colonies identified as E. coli were further characterised for the presence of stx1, stx2, eae, ehxA, rfb O111 and rfb O157 by PCR as described above. Patient ages were recorded for all STEC-positive cases. For patients with STEC O157 or O111 infection, data on hospitalisation, clinical evolution, focused on the development of HUS and outcome were also recorded. The AMR profile of O157 and O111 isolates was determined using the Vitek 2 system (BioMérieux) and the AST-N244 (Gram-negatives) card according to the manufacturer's instructions and clinical cut-off values were applied according to CLSI guidelines [27].
Statistical analysis
Herd-level prevalence estimates were expressed as the percentage of herds/flocks that tested positive in each farm system out of all herds/flocks that were examined in the respective farm system, with 95% confidence intervals adjusted for the population size, using the software EpiInfo2. Differences in prevalence frequencies were assessed using statistical software Stata/IC version 13.1 (StataCorp LP, College Station, TX, USA). Association between variables of interest and proportion of ruminant herds shedding STEC and E. coli O157:H7 was assessed. Variables tested were: (i) farm management system (sheep, beef cattle and dairy cattle), (ii) animal species (sheep and cattle), (iii) sampling season (categorised according to calendar year, i.e. spring, summer, autumn, winter), (iv) geographical location of the farm (oceanic, continental) and (v) herd size stratified according to farm system management as follows: beef cattle,<50, 50–100 and>100; dairy cattle,<50, 50–150 and>150; sheep,<150, 150–300 and>300. Analyses were first performed in the whole dataset to test the association of each of the variables with STEC and E. coli O157:H7 farm prevalence. Then, data were stratified to the variables associated with statistically significant differences. Annual incidence rate of human clinical cases was calculated as the number of cases accumulated in the 3 years for each age group divided by the population size at risk (i.e. population size for each age group multiplied by 3 years at risk of event), and then referred to 100 000 inhabitants. The effect of seasonality in E. coli O157:H7 and non-O157 human infections was evaluated with data stratified by age (infants, 0–5 years; teenagers, >5–15 years; adults, >15–65 years; elderly, >65 years). The distribution of the different virulence genes and profiles of O157 and non-O157 STEC isolates from different sources was compared. Thus, virulence genes distribution (presence or absence) was compared between isolates from animal–human, cattle–human, sheep–human, cattle–sheep and dairy cattle–beef cattle. For virulence genes profiles, variables were stratified to two categories (the category of interest and all other categories) and its presence compared between isolates of animal and human origin. One sample test of proportions with 95% CI was performed to look for differences in eae gene carriage among non-O157 human clinical isolates. The χ 2 or Fisher's exact tests were run, depending on sample size and expected frequencies [Reference Cochran28]. In all cases (animal and human), when overall significance was < 0.05, post-hoc analysis involving pairwise comparisons of two proportions was performed. Bonferroni correction was applied to compensate for multiple groups testing (to control Type I error) and statistically significant differences were determined based on adjusted P-values (P-value 0.05 divided by the total number of pairwise comparisons).
RESULTS
Prevalence of STEC-positive herds/flocks
The distribution of herds positive to STEC (any serotype) and E. coli O157:H7 by host animal species is shown in Table 1. STEC were more frequently isolated from beef cattle (63.5% of herds) and sheep (56.5% of flocks) than from dairy cattle (30.5% of herds) (χ 2 = 13.07, P < 0.001 and χ 2 = 19.95, P < 0.001, respectively). STEC serogroup O157 were only isolated when IMS was performed, and all isolates identified as E. coli O157 carried the H7 flagellar antigen gene fliC, thus confirming the strains to be serotype O157:H7. STEC serogroup O157 were more frequently isolated from sheep flocks (20.0%) and beef cattle herds (18.3%) than from dairy cattle herds (14.6%) but differences were non-significant (P = 0.623). Only in the case of sheep flocks, differences in E. coli O157:H7 prevalence were found between sampling seasons (χ 2 = 8.08, P = 0.044); prevalence in summer and autumn being higher than winter (P = 0.027 and P = 0.016, respectively). However, these differences were no longer significant after Bonferroni correction. STEC serogroup O111 was not detected in this study. Regarding herd size, no significant associations with STEC (any serotype), O157 STEC or non-O157 prevalence (P > 0.05) were observed when considering all farms together. When comparisons were stratified by farm system, only beef cattle showed differences in O157 STEC excretion, with medium-sized herds presenting higher rates of O157 STEC than small farms (χ 2 = 7.361, P = 0.007). No significant associations were found between STEC shedding and any of the other variables tested.
Table 1. Herds/flocks positive to Shiga toxin-producing Escherichia coli (STEC) (any serotype), O157:H7 STEC and non-O157 STEC

Characterisation of virulence genes of STEC isolated from ruminants
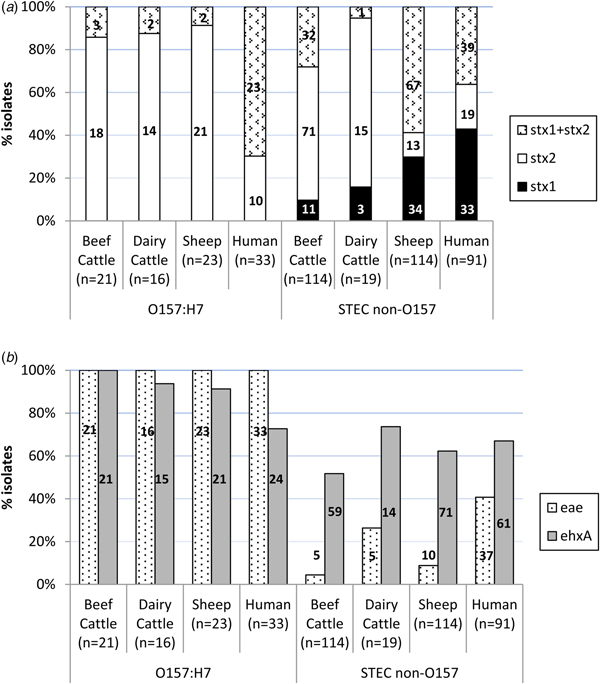
An average of two colonies (maximum six) per STEC-positive herd/flock were characterised for the presence of stx1, stx2, eae and ehxA, amounting to a total of 307 STEC isolates from different sources (135 isolates from 66 beef cattle herds, 35 isolates from 25 dairy cattle herds and 137 isolates from 65 sheep flocks) and serogroups (60 O157:H7 and 247 non-O157). Eleven different patterns of virulence gene combinations were found in STEC from ruminants (Table 2). However, only three profiles were observed among the O157:H7 isolates, with the toxigenic profile stx2 + eae + ehxA as the most prevalent (50/60, 83.3%), but very uncommon (4/247, 1.6%) among non-O157 STEC (Table 2). All E. coli O157:H7 strains carried the stx2 and eae genes, 95.0% (55/60) also carried the ehxA gene and 11.7% (7/60) carried the stx1 gene (always in combination with stx2) (Fig. 1). No differences were found regarding the prevalence of genes in E. coli O157:H7 isolated in the different animal species analysed (P > 0.05). Conversely, the distribution of Shiga toxin genes was clearly different in non-O157 isolates collected from sheep and cattle (Table 2, Fig. 1A). Hence, stx2 gene alone was rarely found in sheep isolates (13/114, 11.4%), which more frequently carried stx1 and stx2 genes (67/114, 58.8%); whereas in cattle, stx2 gene alone was more prevalent (86/133, 64.7%). All comparisons of Shiga toxin gene distribution in non-O157 STEC between cattle and sheep were statistically different (P < 0.001). Presence of eae and ehxA genes among non-O157 isolates was not as widespread as in O157:H7 isolates (Fig. 1B), and no differences were found between sheep and cattle (P > 0.05) but eae was more prevalent in isolates from dairy than beef cattle (P = 0.006). The combination of eae and ehxA genes in non-O157 isolates from ruminants was very rare (13/247, 5.3%). Herds shedding non-O157 STEC-positive for any stx gene (stx1 and/or stx2) and eae gene would include four beef cattle herds (3.8%; five isolates), five dairy cattle herds (6.1%; five isolates) and eight sheep flocks (7.0%; 10 isolates). If only the more pathogenic combination stx2 + eae was considered, the percentage of herds shedding this particular non-O157 STEC genotype would be even smaller (3.8% of beef cattle herds, 3.6% of dairy cattle and 5.2% of sheep flocks).
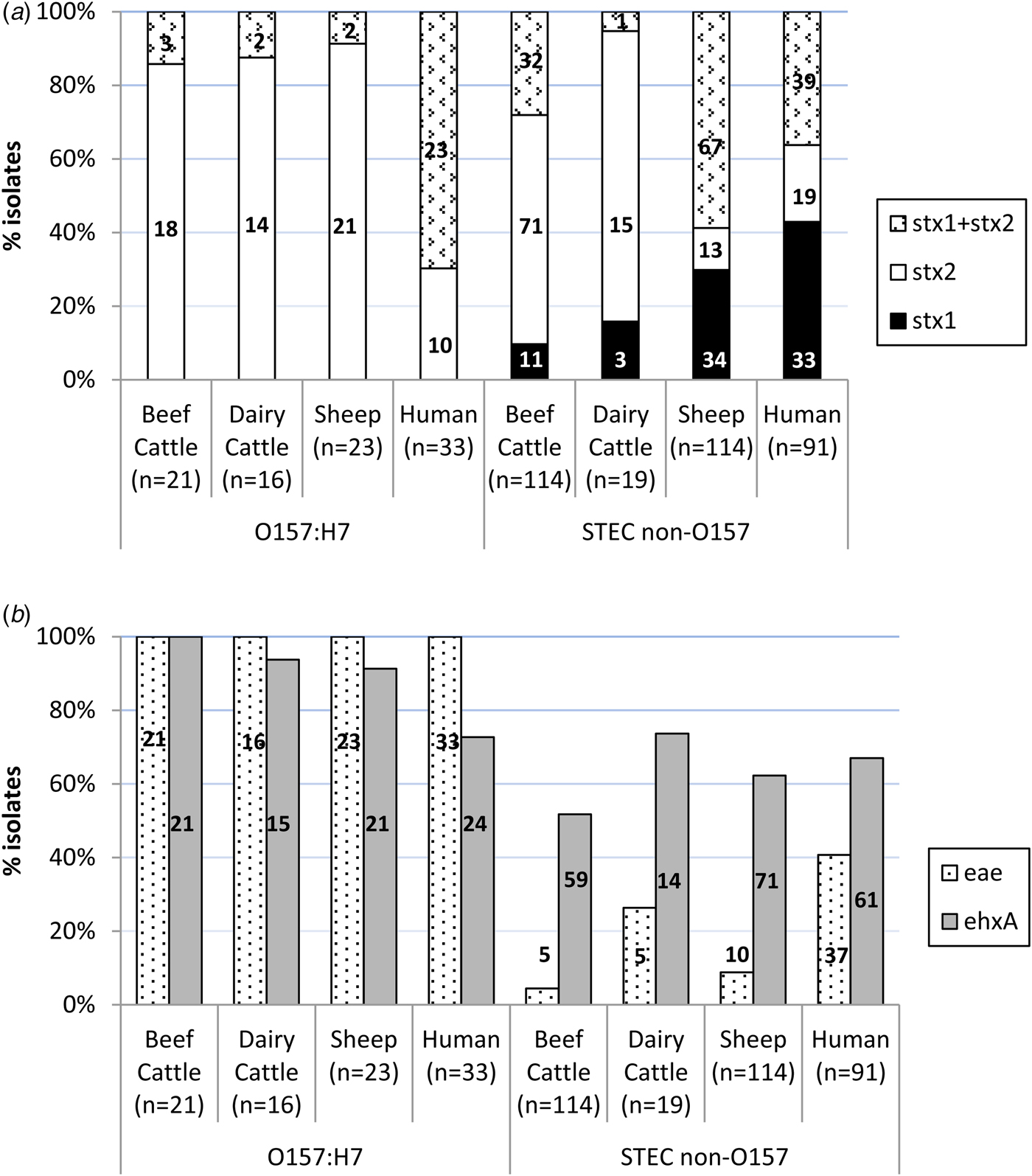
Fig. 1. Distribution of virulence genes among Shiga toxin-producing Escherichia coli from ruminants (307 isolates from 156 herds) and human clinical cases (124 isolates). (A) Shiga-toxin genes; (B) eae and ehxA genes. Number of isolates is indicated within the columns.
Table 2. Combination of virulence genes among Shiga toxin-producing Escherichia coli from ruminants (307 isolates from 156 herds) and human clinical cases (124 isolates)

Prevalence of STEC in human patients
Of the 29 094 stool samples screened by multiplex-PCR, 233 were stx-positive. Culture of the 233 stx-positive stool samples yielded 162 isolates confirmed by MALDI-TOFF as E. coli, and 124 of them were identified as STEC by individual colony PCR targeting the stx1 and stx2 genes. Of the 124 STEC, 33 were identified as belonging to serogroup O157, four to serogroup O111 and the remaining 87 isolates belonged to other undetermined non-O157 serogroups. Thus, O157:H7 STEC accounted for 0.11% (33/29 094) cases of gastrointestinal infection and non-O157 STEC for 0.31% (91/29 094). Mean annual incidence rate calculated for the population served by the hospital was 1.7 cases/100 000 inhabitants for O157 STEC and 4.7 cases/100 000 inhabitants for non-O157 STEC, but cases mainly concentrated among young patients (Fig. 2). Actually, of the 33 patients with O157 STEC detected in stool samples, 17 were <12 years old. STEC infections showed a seasonal distribution, with the largest proportion of STEC cases occurring in summer, followed by autumn. In fact, O157 STEC cases concentrated in summer and non-O157 cases in autumn. With stratification by age, these seasonal differences in STEC carriage (both O157 and non-O157) were observed in infants and teenagers (Fisher's exact test, P = 0.007 and P = 0.023, respectively). However, after pairwise comparisons between seasons, differences were only observed for infants, prevalence of O157 infections in summer being significantly higher than in autumn (χ 2 = 8.044, P = 0.005).

Fig. 2. Annual incidence of confirmed STEC (O157 and non-O157) human infections by age.
The main clinical symptoms included diarrhoea accompanied with vomiting or abdominal pain; in addition, 11 patients had bloody diarrhoea. Twelve patients were admitted to the hospital with a suspicion of gastroenteritis. A 9-year-old boy with stool sample positive for O157 STEC developed HUS that resolved without any sequelae. Another HUS case was observed in a 2-year-old child due to E. coli O111 who also recovered without sequelae. Two male patients, aged 57 and 79 years, infected with O157:H7 E. coli died; both patients were immunocompromised and had comorbidities.
Characterisation of virulence genes of STEC isolated from human patients
Distribution of stx1, stx2, eae and ehxA in the 124 STEC isolates of human origin is shown in Table 2 and Figure 1. All 12 possible patterns of virulence genes combinations were found in human STEC isolates; these included the 11 virulence gene patterns that had been found in animals plus an additional pattern (stx1 + stx2 + eae) found in both O157 and non-O157 human isolates but not in animals. In human O157:H7 isolates, stx1 + stx2 + eae + ehxA was the most common virulence gene profile (15/33, 45.5%) found, followed by stx1 + stx2 + ehxA (27.3%, 9/33). All E. coli O157:H7 strains from human clinical cases carried the stx2 and eae genes, 72.7% (24/33) carried the ehxA gene and 69.7% (23/33) the stx1 gene. However, human O157:H7 isolates more frequently carried stx1 and stx2 genes together (23/33, 69.7%) than stx2 gene alone (10/33, 30.3%); stx1 alone was not observed. Among non-O157 isolates, stx1 alone (39/91, 42.9%) was more prevalent than stx1 + stx2 (33/91, 36.3%) or stx2 alone (19/91, 20.9%); ehxA was present in 67.0% of the cases (61/91) and eae in 40.7% (37/91). Overall, non-O157 human isolates were more likely to be eae-negative than eae-positive (P = 0.037). When stratified by age, this difference was only significant for adults (>15–65 years, P = 0.005) and the elderly (>65 years, P = 0.006). The combination stx2 + eae was found in 16.5% (15/91) of non-O157 human clinical isolates. The four serogroup O111 isolates recovered had the following profiles: stx1 + stx2 + eae + ehxA (two isolates), stx2 + eae + ehxA (one isolate) and stx1 + eae (one isolate).
Comparison of virulence profiles between animal and human STEC
Even though all but one of the virulence profiles described in this study (Table 2) were found in both animals and humans, there were statistically significant differences in the prevalence of virulence genes and profiles in animals and humans. Although in E. coli O157:H7 strains, the genes stx2 and eae were present in all isolates from both ruminants and humans, stx2 alone was more prevalent in ruminant than in human isolates (P < 0.001) while the combination of stx1 + stx2 was more frequent in humans (P < 0.001). The ehxA gene was significantly more prevalent among E. coli O157:H7 isolates from animals than humans (P = 0.004). Whereas in animal isolates the gene profile stx2 + eae + ehxA was the most prevalent among serotype O157:H7, in human isolates stx1 + stx2 + eae + ehxA was the most commonly found. Regarding non-O157 STEC, gene distribution was clearly different between ruminant and human isolates. Hence, the presence of the gene stx2 alone was again significantly associated with animal isolates (P = 0.001), mainly due to its widespread distribution in cattle, while stx1 alone (P < 0.001) was significantly associated with human isolates; the combination stx1 + stx2 did not significantly differ between human and animal isolates considered globally (P = 0.526), but it was significantly different when comparing human and sheep isolates (P = 0.002). The eae gene was significantly associated with human isolates (P < 0.001), and the ehxA gene was, however, more evenly distributed among the different isolation sources (P = 0.115). The highly pathogenic profile stx1 + stx2 + eae + ehxA was more frequently associated with non-O157 human clinical isolates (P = 0.003), as were two other gene combinations containing stx1 gene (stx1 + eae + ehxA, P < 0.001; stx1 + ehxA, P = 0.011). Another three virulence gene profiles were more frequently associated with animal isolates (stx2, P = 0.018; stx2 + ehxA, P = 0.040; stx1 + stx2 + ehxA, P = 0.003).
AMR profiles of STEC isolated from ruminants and humans
AMR profiles were determined for 106 STEC isolates of animal origin (58 O157 and 48 non-O157 isolates representing the 11 virulence genes profiles) and 36 isolates from human clinical cases (32 O157 and four O111). Antimicrobials tested, distribution of MICs and interpretation of results are shown in Table 3. Most of the animal isolates (103/106, 97.2%) were susceptible to all 14 antibiotics tested; only three O157:H7 isolates (two from sheep and one from dairy cattle; gene profiles stx2 + eae + hlyA – one isolate, and stx1 + stx2 + eae + hlyA – two isolates) showed a multiple drug resistance (MDR) profile with resistance to ampicillin, trimethoprim, sulfamethoxazole and tetracycline (Table 4). In the case of human isolates, 80.6% (29/36) were susceptible to all 16 antimicrobials tested, including the two isolates obtained from the patients who developed HUS and the two deceased patients. Seven isolates (all of serogroup O157) were resistant to at least one antimicrobial, and the highest percentage of isolates were resistant to trimethoprim/sulfamethoxazole (16.7%) followed by ampicillin (13.9%). Multidrug resistance phenotypes included resistance to two, three and six antimicrobials (Table 4).
Table 3. Resistance (percentage) and distribution of MICs for the 106 ruminant STEC and 36 human STEC (O111/O157) isolates

White fields denote range of dilutions tested for each antimicrobial agent. MICs equal to or above the range are given as the concentration closest to the range and indicated in bold. MICs equal to or lower than the lowest concentration tested are given as the lowest tested concentration. Vertical lines indicate cut-off values: EUCAST epidemiological cut-offs (for ruminant isolates) are represented by thicker lines; CLSI clinical cut-offs (for human isolates) in thin lines, dashed for intermediate and continued for resistant.
aNo cut-off value given by EUCAST; reference as indicated by double vertical lines were used.
bTrimethoprim/sulfamethoxazole cut-off values are expressed as trimethoprim concentration (range 1:19–16:304).
cAll resistant isolates belonged to serotype O157:H7.
dIntermediate resistance.
Table 4. Resistance phenotypes of STEC isolates

a AMP, ampicillin; AMC, amoxicillin/clavulanic acid; GEN, gentamicin; NAL, nalidixic acid; SXT, sulfamethoxazole/trimethoprim (tested separately in animal isolates); TBM, tobramycin; TET, tetracycline.
b Intermediate resistance to AMC and TBM.
DISCUSSION
In this study, we present an updated prevalence estimate of STEC in the main animal reservoirs in the Basque Country along with parallel data on human clinical cases, to contextualise the current risk that ruminants represent to human health and to monitor AMR in the context of a one health approach. The animal survey included not only beef cattle but also dairy cattle and sheep, the latter an important production system in the region for reasons related with traditional and artisan cheese production under Protected Designation of Origin (PDO). Accurate estimates of herd prevalence at each production system were obtained and a collection of strains representative of the region was compiled for characterisation. Data from human clinical cases were obtained from one of the main hospitals of the Basque Country to assure good reporting coverage while collecting a representative number of isolates.
The results of the cross-sectional survey carried out in ruminants showed a high proportion of STEC-positive cattle and sheep herds, at levels similar to those reported in a comparable study carried out in the region in 2003–2005 [Reference Oporto12]. Interestingly, and in agreement with the results found in that previous study, the proportion of herds shedding STEC was higher in semi-intensive management systems (beef cattle and sheep) compared with intensive systems (dairy cattle); a similar trend was also observed for O157:H7 STEC. Other authors also found an association between pasture access and STEC shedding [Reference Venegas-Vargas29], but opposite results have also been reported [Reference Cobbaut30, Reference Cobbold31]. Animal management or other practices specific to livestock production in different countries might explain these discrepancies [Reference Callaway32]. In addition, herd size has also been shown to affect infection prevalence when a higher number of susceptible hosts are continuously challenged by contact with carriers [Reference Huston33, Reference Davison34]. Here, the effect of herd size was only significant in beef cattle, with medium-sized herds presenting higher rates of O157 STEC than small farms, but reasons are unknown. When considering herds shedding E. coli O157:H7, prevalence was higher than that reported in our previous study carried out 10 years earlier, which was estimated to be 8.7% for sheep flocks and 3.8% for cattle herds [Reference Oporto12]. However, results from that study were probably an underestimation of the true prevalence due to the methodology used. Still this increasing trend observed in ruminants would be in accordance to the epidemiology of enterohaemorrhagic E. coli infections in humans in the Basque Country, which rose from 0.28 to 1.11 cases/100 000 inhabitants in 12 years (2004–2016) [13, 15]. Similarly to the data obtained for animals, the data collected at the Hospital Universitario Donostia also showed an increasing trend in STEC incidence, but again, improvements in the methodology used could be one of the main reasons for this apparent increase. In agreement with other studies [Reference Griffin and Tauxe35], STEC-associated diarrhoea and HUS cases concentrated among young patients, both for O157:H7 and other non-O157 STEC infections. During the period of this study, two cases of HUS were registered, one caused by E. coli O157 in a 9-year-old boy and another caused by E. coli O111 in a 2-year-old child, both with a favourable outcome. Serogroup O111 has been one of the main causes of STEC outbreaks in the region [Reference Mora10]; however, it was only isolated in four patients in this study. Also, this serogroup seems to be uncommon in ruminants [Reference Mora10, Reference Monagan36], and, in accordance, no O111-shedding herds were found here. The seasonality of STEC described here, with a larger proportion of cases identified during summer and autumn, agrees with other studies [Reference van Duynhoven37–39].
A total of 307 isolates from ruminants were characterised, including 135 from beef cattle, a number that further exceeds those analysed in a nationwide sampling (8 O157 and 56 non-O157) performed in beef cattle in 34 slaughterhouses in Spain in 2011 and 2013 in compliance with Directive 99/2003/EC [Reference Cabal40]. An additional strength of this study is that it also included isolates from dairy cattle and sheep, as well as human isolates collected in the same region, allowing comparison of strains from different sources. Interestingly, the virulence gene profiles in E. coli O157:H7 isolates, but mainly in non-O157 isolates, were different depending on the isolation host. These could be due to the diversity of non-O157 serotypes isolated from different hosts, but indicates that despite the high proportion of STEC shedding herds, only a small proportion of STEC carried by ruminants could be epidemiologically related to clinical disease in humans. Moreover, despite the wide distribution of genes stx2 and eae in O157 STEC, a gene combination considered to be more consistently associated with severe disease in humans [Reference Boerlin41], more than half of patients infected with O157 STEC showed only mild gastrointestinal symptoms. In non-O157 STEC, the stx2 gene was more prevalent in ruminant strains (80.6%), particularly in cattle (89.5%), than in human strains (57.1%), but the eae gene was present in only 8.1% of the ruminant non-O157 strains, compared with 40.6% of those from human clinical cases. This low prevalence of eae-positive isolates among non-O157 from ruminants is in accordance to that found 10 years before [Reference Oporto12] and with several other studies [Reference Monagan36, Reference Urdahl42, Reference Ennis, McDowell and Bolton43], but much lower than the 64.3% recently reported in beef cattle in Spain [Reference Cabal40]. Although this might suggest that only a small proportion of STEC shedding herds pose a high risk, results from human clinical cases reported here and elsewhere [Reference Krause, Barth and Schmidt44] showed that isolates also lacking the eae gene can cause gastrointestinal disease. The fact that a significantly higher proportion of non-O157 human isolates were eae-negative than eae-positive, particularly in adults and the elderly, might indicate that these patients were more frequently infected with serotypes other than the top five non-O157 STEC [45]. Still, all STEC shedding herds should be considered as a potential risk for human infection.
Treatment of human STEC infections is generally supportive and antibiotics are usually contraindicated. However, monitoring AMR patterns of intestinal STEC from healthy ruminants in their intestine and their comparison with those isolated from human clinical cases, provide valuable information of the public health risk posed by animal reservoirs with respect to the transmission of resistant E. coli strains to humans and the spread of AMR to other enteric bacteria. Here, the proportion of resistant ruminant isolates was lower than in other studies [Reference Cabal40, Reference Ennis, McDowell and Bolton43, Reference Mora46], and only found in O157 STEC. However, all resistant isolates exhibited the same MDR profile (ampicillin/trimethoprim/sulfamethoxazole/tetracycline), consistent with the fact that β-lactams, trimethoprim/sulphonamides and tetracyclines are among the most commonly used antibiotics in veterinary medicine [47]. In humans, most isolates tested were O157 strains and 21.9% (7/32) of them showed some level of resistance. Ampicillin resistance was also widespread among human isolates, and all but one of the human O157 isolates with a resistant phenotype were resistant to trimethoprim/sulfamethoxazole. This drug combination is recommended for treating a range of human infections, including infectious diarrhoea when necessary and urinary tract infections (acute uncomplicated cystitis and pyelonephritis) [Reference Gupta48], and therefore dissemination of trimethoprim/sulfamethoxazole resistance in E. coli isolates should be closely monitored. However, all the isolates were susceptible to fluoroquinolones and third-generation cephalosporins, alternative antimicrobials for treating infections caused by sulfamethoxazole/trimethoprim-resistant E. coli. Similarly, all ruminant isolates were susceptible to fluoroquinolones, macrolides, aminoglycosides, third-generation cephalosporins, carbapenems or colistin.
In conclusion, this study represents a thorough survey of STEC infection in ruminants and humans that provided reliable estimates on herd-prevalence data and human incidence rates, and allowed comparison of isolates from different sources. Results showed a wide distribution of STEC in ruminants in the Basque Country, but distribution of virulence factors among STEC strains from sheep and cattle differed from those of human clinical cases. The proportion of resistant isolates in ruminants was not high, but the MDR profiles were similar to those found in human isolates. Therefore, presence of STEC in healthy cattle and sheep represents a potential risk for public health that cannot be neglected.
Author ORCIDs
M. Ocejo, 0000-0001-5136-956X; A. Hurtado, 0000-0001-7089-9670
Acknowledgements
The authors would like to thank the veterinary staff from the Diputaciones Forales de Gipuzkoa, Bizkaia and Araba, and the farmers for their collaboration. The authors thank Dr A.P. Tedim (NEIKER) for helpful comments on the manuscript.
Financial support
This work was funded by the Basque Government. M.O. is the recipient of a predoctoral fellowship from the Basque Government (Departamento de Desarrollo Económico e Infraestructuras).
Conflict of interest
None.