Clostridium difficile is a gram-positive, anaerobic, spore-forming bacterium; it is a leading cause of healthcare-associated infective diarrhea. Risk factors for the disease include previous hospitalizations, advanced age, and the use of antibiotics.Reference Slimings and Riley 1 – Reference Vardakas, Trigkidis, Boukouvala and Falagas 3 Clostridium difficile spores play an important role in disease transmission. Spores can persist in the environment for several months and are resistant to stresses such as heat, oxygen, and disinfectants.Reference Kramer, Schwebke and Kampf 4 , Reference Sethi, Al‐Nassir, Nerandzic, Bobulsky and Donskey 5 Clostridium difficile spores are thought to be ubiquitous in the hospital environment,Reference Samore, Venkataraman, DeGirolami, Arbeit and Karchmer 6 and contaminated hospital surfaces may serve as an important reservoir for transmission.Reference Weber, Rutala, Miller, Huslage and Sickbert-Bennett 7
However, optimal and standardized sampling and laboratory methods for assessing density of environmental C. difficile are not known. Increasing surface area may maximize the potential to capture spores and push C. difficile quantities over the detection limit of a given laboratory method, enabling more precise measurements,Reference Hospodsky, Yamamoto and Peccia 8 , Reference Shams, Rose and Edwards 9 but it might also increase the concentration of inhibitors and reduce detection. Larger surface areas also require larger sampling devices, and sponges are preferred over swabs for sampling such areas due to their larger size.Reference Ali, Muzslay and Wilson 10 , Reference Engelhardt, Foster, Hong, Riley and McGechie 11
Other challenges include how to consistently measure average environmental burden across surface types (eg, bedrails, floors, and other surfaces), and whether culture or culture-independent methods yield more reliable quantitative measures of the density of environmental C. difficile. Bedrails represent one of the most frequently touched surfaces in the hospital environment.Reference Huslage, Rutala, Sickbert-Bennett and Weber 12 Floors, while not a high-touch environment, are often the most C. difficile-contaminated surfaces in hospitals; thus, they may play a role in transmission.Reference Rashid, Vonville, Hasan and Garey 13 While culture techniques may be most readily linked to potential transmission and infection, culture-independent techniques may nevertheless improve quantification of the existing spore burden and risk.Reference Hospodsky, Yamamoto and Peccia 8
In this study, we sought to identify whether increased sample surface area was associated with increased detection of C. difficile from environmental samples collected from patient rooms in a tertiary-care hospital in a nonoutbreak setting. We explored whether this relationship existed for both floor and bedrail samples, and across several different microbiologic methods, including qPCR and enrichment culture.
Methods
Design and setting
We used an efficient split-plot sampling designReference Altman and Krzywinski 14 , Reference Jones and Nachtsheim 15 to assess the impact of sample surface area on C. difficile detection from hospital environmental samples while controlling for room-level clustering of bacterial burden. Small surface-area and large surface-area pairs of bedrail and floor environmental samples were selected from 12 rooms (N=48) in 3 inpatient units (2 general medicine, 1 intensive care), over the course of 2 days in September 2017 in a large tertiary-care hospital located in Toronto, Canada.
Sample collection
For sampling, patient rooms were selected based on a categorization of risk of contamination: high-risk rooms were defined as those of patients with active C. difficile diarrhea identified by hospital infection control staff; medium-risk rooms were those of patients with a history of in-hospital antibiotic use in the last 14 days; and low-risk rooms were those of patients without a history of C. difficile or in-hospital antibiotic use in the previous 14 days. All high-risk rooms available in the hospital were selected (N=5), as well as a selection of medium-risk (N=2) and low-risk rooms (N=5) on the same wards. All samples were collected independently of ward- and room-cleaning schedules, and the timing of most recent cleaning was not known or recorded. The hospital environmental cleaning policy specified daily routine cleaning and terminal cleaning as surface cleaning with quaternary ammonium. Clostridium difficile cleaning was surface cleaning with accelerated hydrogen peroxide performed twice daily. 16
In each selected room, 2 pairs of samples were collected using sterile cellulose sponges premoistened in Dey-Engley neutralizing buffer (Scigiene, Scarborough, ON). The first pair consisted of a small surface-area floor sample (32×32 cm, 0·10 m2) and a large surface-area floor sample (100×100 cm, 1 m2). These samples were collected from contiguous spaces alongside the patient bed on the side of usual access. The second pair consisted of a small surface-area bedrail sample (7·7×30 cm, 0·023 m2) and a large surface-area bedrail sample (77×30 cm, 0·23 m2). These samples extended down the side of the bedrail panel. Again, these samples were contiguous and from the bedrail on the side of usual access. Small surface-area samples were taken from the portion of the bedrail corresponding to the head of the bed, while large surface-area samples included the remainder of the bedrail. Surfaces were wiped using perpendicular, overlapping “S” patterns using a defined protocol. 17
Outcome measurements
Clostridium difficile cells were extracted from sponge swabs. Briefly, 40 mL sterile double-distilled water was added to the sample bag, which was then vigorously massaged between fingers for 90 seconds. Extracted cells and environmental debris were collected from the liquid by centrifugation at 7,500×g for 15 minutes. All but 1 mL of the supernatant was removed, and the pellet was resuspended and transferred to a microcentrifuge tube, which was centrifuged at 10,000×g for 5 minutes. The entire supernatant was removed, and the pellet was resuspended in 500 µL of 20% Dey-Engley neutralizing broth (Scigiene) diluted in sterile water.
With half of the sample, DNA was extracted using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, Irvine, CA) using a 1-hour bead-beating step and a DNA elution volume of 50 µL. We conducted quantitative polymerase chain reaction (qPCR) on 2 targets. A 157-bp conserved region of the C. difficile 16S rRNA gene (present in ~10 copies per genome) and an internal TaqMan probe (Applied Biosystems, Foster City, CA) were used for detection of all C. difficile strains.Reference Mutters, Nonnenmacher, Susin, Albrecht, Kropatsch and Schumacher 18 A 127-bp region of the toxin B gene (present in 1 copy per genome) and a corresponding TaqMan probe were used to specifically detect toxigenic C. difficile strains.Reference Kilic, Alam and Tisdel 19 Reactions (10 µL) containing 5 µL JumpStart Taq ReadyMix for qPCR (D7440, Sigma-Aldrich, St Louis, MO), 0·1 µL reference dye for qPCR (R4526, Sigma), 0·4 µL 25mM MgCl2, 0·5µM (each) forward and reverse primers, 0·15µM TaqMan probe, and 3·85 µL DNA were prepared. These were run in triplicate on an ABI 7900HT thermocycler (Applied Biosystems) under the following conditions: 50°C for 2 minutes, 95°C for 10 minutes, 45 cycles at 95°C for 15 seconds, 60°C for 1 minute. The other half of the sample was enriched and cultured using banana broth (Hardy Diagnostics, Santa Maria, CA).Reference Kilic, Alam and Tisdel 19 Samples positive by enrichment culture were ribotyped (Appendix 1).
We coded 5 outcome variables. First, 16S qPCR positivity and toxin B qPCR positivity were coded such that any threshold cycle (Ct)<45 was considered positive, and any Ct ≥ 45 (or undetermined) was considered negative. The enrichment cultured-based measure was coded as either positive or negative. Second, we converted qPCR Ct to spore counts for 16S qPCR and for toxin B qPCR based on their respective standard curves. These C. difficile standard curves were generated by serial dilution of a spore stock quantitated using a hemocytometer. As is customary in occupational exposure assessment analyses, we truncated the estimated spore count distribution at 0·5 spores, corresponding to half the detection limit.Reference Ogden 20
Covariates
We coded risk-factor variables corresponding to sample type (bedrail or floor) and room risk of contamination (low, medium, and high). Sample surface area was encoded as a categorical variable (small or large). To adjust for surface area in our multivariableReference Hidalgo and Goodman 21 models, we also encoded sample surface area in as a continuous, log10-transformed variable, log10 (m2). Finally, we also coded 3 clustering variables: sample pair identifier, room identifier, and ward identifier.
Statistical analyses
For descriptive analyses, we measured the proportion of samples that were positive for the binary outcome variables and the geometric (log10) mean spore count for the continuous outcome variables across the 3 risk-factor variables.
For multivariable analyses, we used separate logistic mixed effects models for the presence versus absence of C. difficile by 16S qPCR, toxin B qPCR, and enrichment culture. We used negative binomial mixed effects models for the estimated spore count by 16S qPCR and toxin B qPCR.Reference Guthrie, Gammill and Kamper-Jørgensen 22 All multivariable models included the same fixed effects and random intercepts. We used fixed effects for patient risk stratum, sample type, and sample surface area, and we used random intercepts corresponding to the ward, the patient room, and the sample pair. The random intercepts for patient room and sample pair accounted for the clustering inherent to the split-plot sampling design.Reference Altman and Krzywinski 14 , Reference Jones and Nachtsheim 15 All multilevel models were fit using Bayesian random-effects regression models in the R statistical programming language (rstanarm library) and default settings (4 chains of 1,000 warmup and 1,000 sampling draws, and default weak priors 23 ). Furthermore, 95% confidence intervals (95% CIs) were based on the sampling draws.
We conducted 2 secondary analyses. First, to assess whether the association between sample surface area and yield was consistent across the surface type (bedrail vs floor), we ran an additional negative binomial multivariable model that added the interaction term between surface area and type. Second, to assess the relatedness of the 3 different laboratory techniques for measuring C. difficile, we measured the association between 16S qPCR spore counts and toxin B qPCR spore counts using linear regression, and between 16S qPCR spore count and enrichment culture positivity using logistic regression.
Ethics
This study was approved by the Public Health Ontario and Sunnybrook Health Sciences Center research ethics boards.
Results
Of the 48 samples collected, 31 (64·6%) were positive for C. difficile by 16S qPCR, with a geometric mean of 13·8 spores per sample; 19 (39·6%) were positive by toxin B qPCR, with a geometric mean of 1·9 spores per sample; and 21 (43·8%) were positive by enrichment culture.
Descriptive analyses
Large surface-area samples were more likely to be positive and had higher estimated spore counts compared to small surface-area samples (Table 1, Appendix 2). By 16S qPCR, 17 of 24 large surface-area samples (70·8%) were positive, while 14 of 24 small surface-area samples (58·3%) were positive. The geometric mean estimated spore count was 27·0 for large surface-area samples and 7·0 for small surface-area samples. However, large surface-area bedrail samples (0·23 m2) were less likely to be positive (5 of 12, 41·7%) than small surface-area floor samples (0·10 m2; 9 of 12, 75%) (Fig. 1). Overall, 21 of 24 floor samples (87·5%) were positive compared to 10 of 24 bedrail samples (41·7%), and the geometric mean estimated spore count was 69·4 for floor samples compared to 2·7 for bedrail samples.
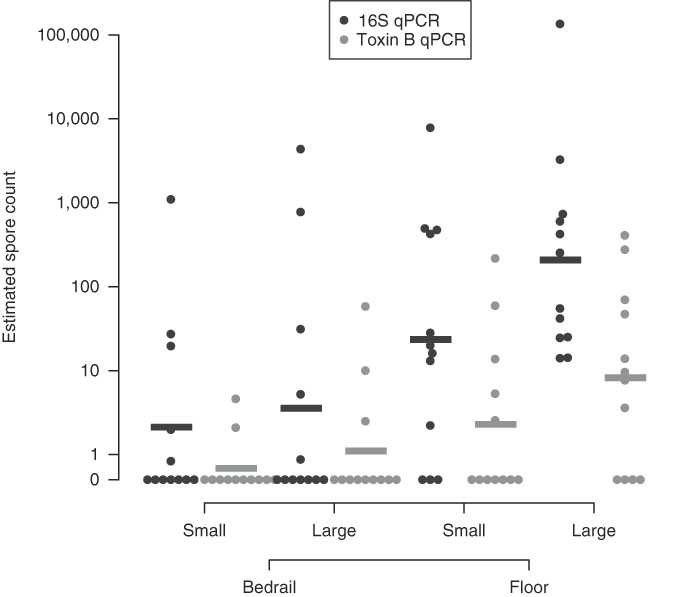
Fig. 1 Estimated C. difficile quantity based on quantitative PCR (16S target and Toxin B target). A horizontal bar is used to indicate the group geometric mean.
Table 1 Estimated Environmental Sample Clostridium difficile Positivity and Quantity Based on qPCR (16S target and Toxin B target) and Enrichment Culture From Environmental Samples Collected at a Tertiary Hospital
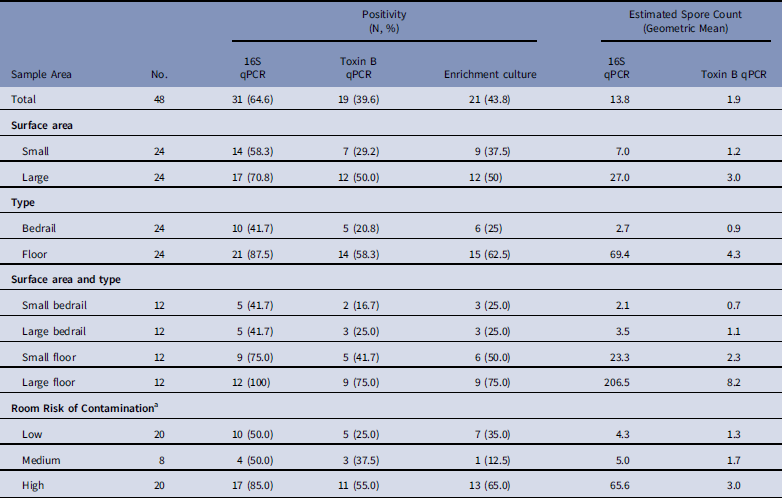
NOTE. qPCR, quantitative polymerase chain reaction.
a Low, occupant had no receipt of antibiotics in prior 14 days; medium, occupant had receipt of antibiotics in prior 14 days; high, occupant had confirmed C. difficile infection.
Multivariable analyses
After multivariable adjustment, each 10-fold increase in surface area was associated with a 4·5-fold (adjusted odds ratio [aOR], 4·5; 95% CI, 0·79–34) increase in the odds of positivity and a 6·6-fold (adjusted count ratio [aCR], 6·6; 95% CI, 3·2–13) increase in the spore count (Table 2). Floor surfaces were 17-fold (OR, 17; 95% CI, 2·1–170) more likely to be positive than bed rails and had 27-fold (aCR, 27; 95% CI, 4·9–181) more spores. Compared to rooms of patients that received no antibiotics in the previous 14 days, samples from rooms of patients with C. difficile infection were 17-fold (aOR, 17; 95% CI, 1·2–246) more likely to be positive, with 11-fold (aCR, 11; 95% CI, 0·55–164) more spores. Models for toxin B qPCR and enrichment culture returned analogous findings. A sensitivity analysis that considered separate surface-area effects for bedrail and floor pairs by 16S qPCR spore count revealed that each 10-fold increase in surface area was associated with a 3·7-fold (95% CI, 1·3–9·8) increase in bedrail spore count and a 9·0-fold (95% CI, 4·2–19·9) increase in floor spore count (P=·08).
Table 2 Predictors of Environmental Sample Clostridium difficile Positivity and Quantity Based on qPCR (16S target and Toxin B target) and Enrichment Culture From Environmental Samples Collected at a Tertiary-Care Hospital, Based on Logistic and Linear Multilevel Regression Models
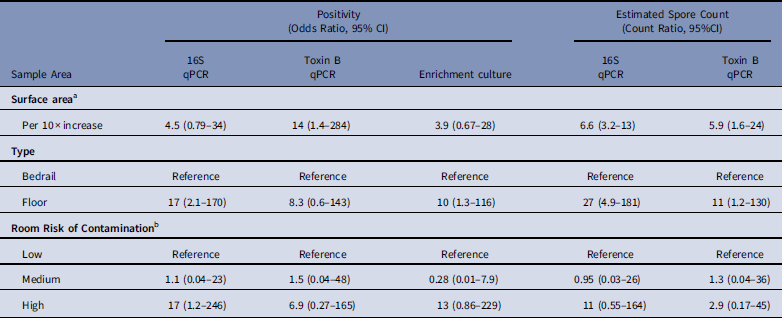
NOTE. qPCR, quantitative polymerase chain reaction; CI, confidence interval.
a For multivariable analyses, sample surface area was coded as log10(m2) so that the effect estimate represents the odds ratio or count ratio per 10-fold increase in the surface area.
b Low, occupant had no receipt of antibiotics in prior 14 days; medium, occupant had receipt of antibiotics in prior 14 days; high, occupant had confirmed C. difficile infection.
Comparison of 16S qPCR versus toxin B qPCR and enrichment culture
Estimated spore counts by 16S qPCR were strongly associated with toxin B spore counts (Pearson’s ρ, 0·75) (Fig. 2, panel A) and with enrichment culture-based results. When the 16S spore count was 0, only 1 of 19 enrichment culture samples (5·3%) were positive. For comparison, 8 of 16 samples (50%) were culture positive for spore counts of 1 to 100; 7 of 8 samples (87·5%) were culture positive for spore counts of 100 to 1000; and 5 of 5 samples (100%) were culture positive for spore counts>1,000 (P LogisticTrend<·001) (Fig. 2, panel B).
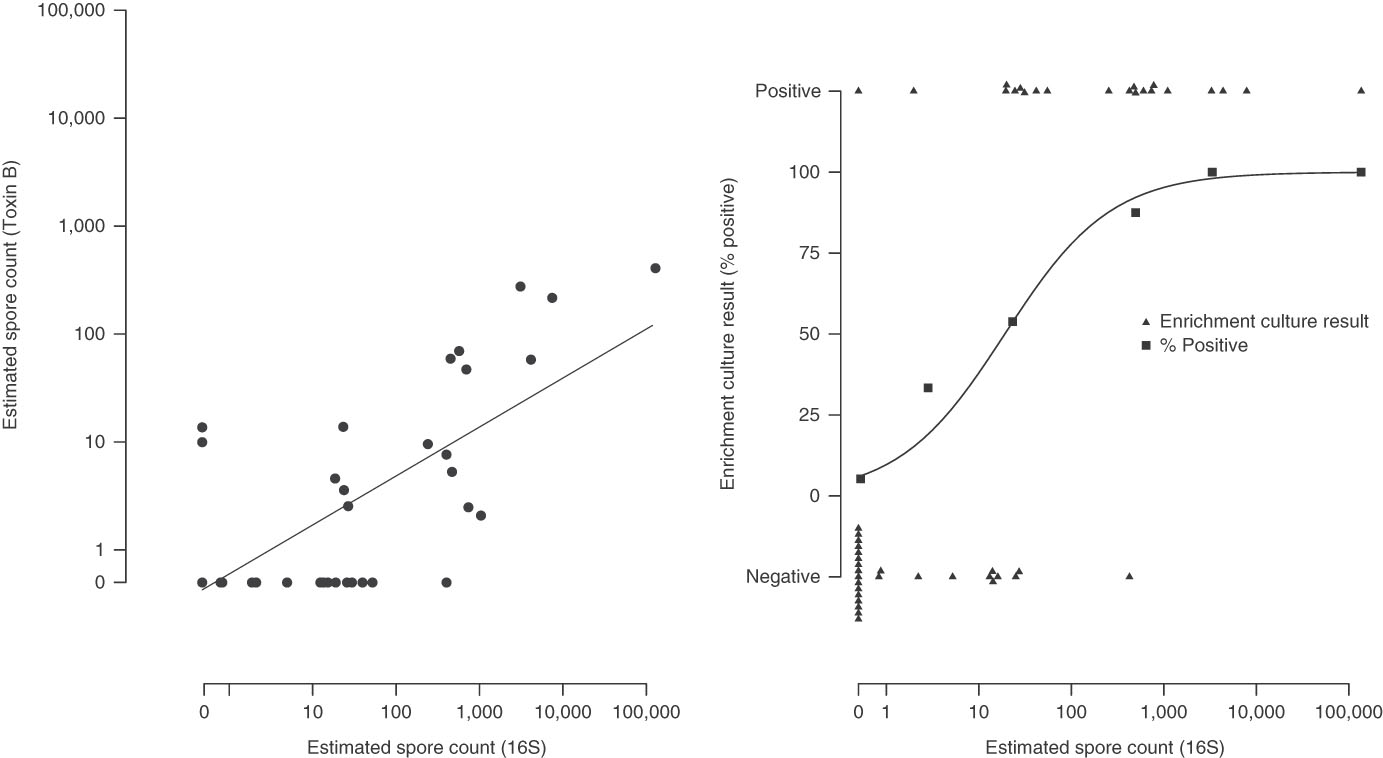
Fig. 2 Association between the testing methods. Panel A: 16S qPCR and Toxin B qPCR. Panel B: 16S qPCR and enrichment culture.
Ribotyping
We successfully ribotyped 20 of 21 samples (95%) that were positive by enrichment culture, and we detected 8 different ribotypes (Table 3). Single ribotypes were identified in 16 of 20 samples (80%), and 2 ribotypes were identified in 4 of 20 samples (20%). Ribotype 015 was found in 11 of 12 positive samples (92%) from ward B, but 0 of 8 samples (0%) were positive from ward A (P χ2=·04). Of 20 samples, 17 (85%) contained ribotypes known to have toxin A and B genes.
Table 3 Characteristics of Clostridium difficile Samples Successfully Ribotyped (N=20)
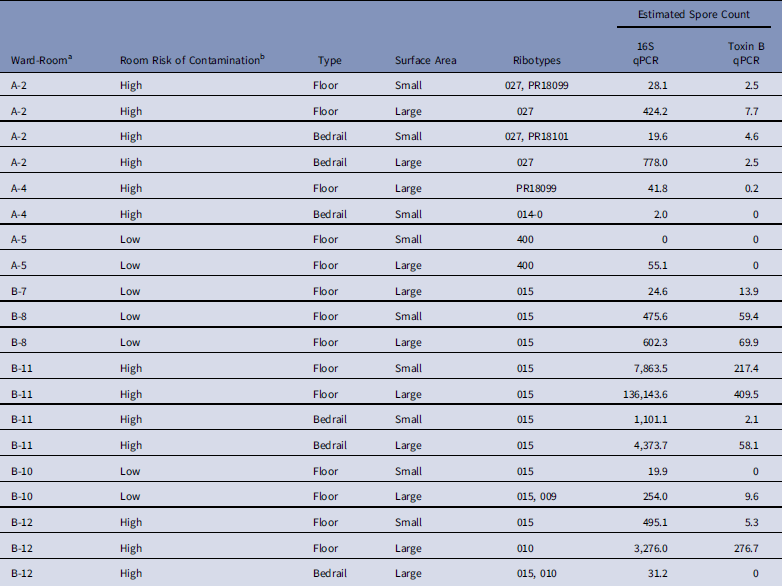
NOTE. qPCR, quantitative polymerase chain reaction.
a No positive samples were obtained from ward C.
b Low, occupant had no receipt of antibiotics in prior 14 days; medium, occupant had receipt of antibiotics in prior 14 days; high, occupant had confirmed C. difficile infection.
Discussion
In this study, we considered the impact of increasing sample surface area on C. difficile recovery using culture and culture-independent techniques. We found that increased sample surface area was associated with increased counts of spores recovered and increased proportions of samples that were positive by all laboratory techniques. Additionally, we identified that 16S qPCR, toxin B qPCR, and enrichment culture are all acceptable techniques for measurement of environmental C. difficile. We also found that C. difficile contamination was common on bedrails and floors and was significantly more common in the rooms of patients with C. difficile infection.
Several techniques have been used to measure environmental C. difficile contamination, including culture,Reference Buggy, Wilson and Fekety 24 enrichment culture,Reference Nerandzic and Donskey 25 and qPCR.Reference Mutters, Nonnenmacher, Susin, Albrecht, Kropatsch and Schumacher 18 Culture-based techniques are commonly used and inexpensive, but they can be plagued by low detection limits, issues of nonculturability, and long turnaround times for results. Although qPCR is a more sensitive and culture-independent technique, it is also more labor intensive and costly, largely due to the DNA extraction step. However, it can produce results within 2–3 hours. qPCR is routinely used for clinical diagnostics and has also been adapted for environmental sampling.Reference Mutters, Nonnenmacher, Susin, Albrecht, Kropatsch and Schumacher 18 We found that 16S qPCR was more sensitive than toxin B qPCR. We also found that results from qPCR-based techniques and culture-based techniques were strongly correlated. Increasing levels of contamination by 16S qPCR were strongly associated with increased levels by toxin B qPCR and enrichment culture-based positivity. At levels beyond 1,000 spores by 16S qPCR, all toxin B qPCR samples and enrichment culture samples were positive, suggesting that toxin B qPCR and enrichment culture may have been negative due to a lack of sensitivity of the laboratory technique rather than a lack of toxigenic or live cells on those surfaces.
Using real-world environmental samples, this study demonstrated that large surface-area samples yielded higher positivity and higher counts of spores across several different microbiologic techniques and 2 surface types. These findings provide further evidence of the advantages of maximizing surface area when conducting environmental sampling. For 16S qPCR, the effect of surface area on C. difficile capture may have been moderately stronger among floor samples than among bedrail samples. This finding may have been due to inconsistencies in the sampling of bedrails because bedrails have complex surfaces. Previous studies have shown that, for Bacillus atrophaeus, large surface-area sampling (1 m2) translates into greater sensitivity for detection and necessitates fewer samples for testing exposure–outcome associations, compared with samples with smaller surface areas.Reference Buttner, Cruz, Stetzenbach, Klima-Comba, Stevens and Emanuel 26 A more recent study used large surface area samples (up to 0·22 m2) to quantify the microbial bioburden of several hospital pathogens.Reference Shams, Rose and Edwards 9 Our work showed that all techniques tested, including both culture and culture-independent techniques, reacted positively to increased sample size, though the clinical relevance of environmental C. difficile burden has not yet been established.
We found that C. difficile contamination was ubiquitous in the hospital rooms we sampled. All rooms, even those of patients without a history of C. difficile or antibiotic use, yielded positive samples by 16S qPCR. It has been shown that floor-surface samples are approximately twice as likely to be culture positive as bedrail samples,Reference Samore, Venkataraman, DeGirolami, Arbeit and Karchmer 6 and surfaces more proximate to patients are more likely to be contaminated than those farther from patients.Reference Moore, Muzslay and Wilson 27 In our study, floor surfaces actually had 10 times more C. difficile contamination than bedrail surfaces. Also, 100% of the 12 large floor samples we collected were contaminated with C. difficile according to 16S qPCR. Clostridium difficile on floor surfaces may be a risk to patients because microbes on the floor may be transported within wards by shoe solesReference Rashid, VonVille, Hasan and Garey 28 are continually resuspended by foot traffic.Reference Qian, Peccia and Ferro 29 Furthermore, mobile patients can contaminate linens and themselves via their feet or shoes.Reference Rashid, Vonville, Hasan and Garey 13 Although our results do not allow us to assess directionality, the finding of strong relatedness of strains recovered within wards suggests that spatial dissemination involving floors may be a concern. These results also lend support to several studies that have shown the importance of ward-level effects for the transmission of C. difficile and other hospital-associated infections.Reference Brown, Valenta, Fisman, Simor and Daneman 30 – Reference Tosas Auguet, Stabler and Betley 32
The infective dose of C. difficile for a healthy but susceptible inpatient is not knownReference Weese 33 and as such, safe levels of environmental C. difficile are not known. For mice, the environmental infectious dose required to infect 50% of mice (ID50) for C. difficile was 1 hour of exposure to 5–10 culturable spores per square centimeter of cage floor.Reference Lawley, Clare and Deakin 34 Future research should seek to identify the infectious dose for C. difficile for both healthy individuals and those at a risk of C. difficile infection due to antibiotic receipt and to define the maximum threshold of environmental density of C. difficile that confers an acceptably low risk of infection. Such environmental density thresholds could be used to better ascertain the acceptability of current hospital cleaning protocols.
Our study has several limitations. First, it is a study of rooms in a single hospital over 2 days of sampling and may not be generalizable to other hospitals. We lacked several covariates including antibiotic history prior to hospital admission and timing of most recent cleaning prior to sample collection. However, these limitations were controlled for by the experimental design of our study, wherein sample surface area and sample type, our 2 exposures of primary interest, were compared to each other within rooms at the same point in time. Second, our small surface-area bedrail samples were selected from the head of the bedrail, while our large surface-area samples were chosen from the remainder of the bedrail, which could have led to a bias if spore density was higher or lower near the head of the bedrail.
In this study, we found that increased sample surface area was associated with increased counts of spores recovered and increased proportions of samples that were positive. We also found that C. difficile contamination was common on hospital bedrails and floors and even more so in the rooms of patients with C. difficile infection. Future studies attempting to quantify the environmental density of C. difficile should consider using large surface-area samples whenever possible. Further study is required to better understand the role of contaminated hospital floors in the transmission of C. difficile infection.
Supplementary materials
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2018.103
Acknowledgments
The authors thank Kwaku Adomako and Arezou Saedi for assistance with research ethics approval, sample collection, and transportation, and Victoria Williams for assistance with coordinating sample collection.
Financial support
This study was funded by a grant from the Canadian Institutes for Health Research. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflicts of interest
All authors declare no conflicts of interest relevant to this article.







