Introduction
There is a well-established shortage of organs available for transplantation in the United States, and demand for suitable organs for transplantation becomes greater every year. Data from the Organ Procurement and Transplantation Network cites ~124,000 Americans currently waiting for an organ transplant. Every day, 22 Americans die waiting for an organ transplant, and one organ donor can save up to 8 lives. Despite a preponderance of organs being transplanted from donors after brain death (DBDs), organ function is not equal to that of living donor organs. Grafts from DBDs have been shown to have inferior outcomes, necessitating repeat transplantation in up to 18% of cases, as compared to less than 10% in living donors [Reference Rao and Ojo1]. Kidney grafts from DBDs have significantly higher delayed graft function (DGF) rates and lower 1-, 3-, and 5-year survival rates, even when compared with living donors with more Human Leukocyte Antigen (HLA) mismatches [Reference Freitas2]. Efforts to improve the management of DBDs before recovery of organs may increase the number of organs transplanted per donor and improve graft function and survival.
After a catastrophic neurologic injury that results in brainstem herniation and the resulting cessation of brain activity, the rest of the body’s organs continue to function, albeit under considerable hemodynamic, endocrine, and immunologic stress [Reference Cooper, Novitzky and Wicomb3, Reference Novitzky, Cooper and Reichart4]. While there have been reports describing protein expression related to brain death (BD)-induced renal injury [Reference Nijboer5, Reference Bouma, Ploeg and Schuurs6], the effect of herniation on inflammatory pathway gene expression in potentially transplantable organs remains incompletely understood. Several groups have described the inflammatory response to brainstem herniation, but these studies are often performed on tissues immediately before implantation, after prolonged periods of both warm and cold ischemia [Reference lznerowicz7]. In transplant recipients, circulating and tissue inflammatory mediators, including selectins, cellular adhesion molecules, and cytokines have been observed at increased levels in humans and animals after receiving an organ from a DBD [Reference Kusaka8–Reference Dziodzio, Biebl and Pratschke10]. Increases in these circulating and tissue mediators in the recipient have been shown to be associated with poor graft outcomes in heart, liver, and renal transplants [Reference Wilhelm11–Reference Damman16]. These studies have looked at small animal models, examined circulating or tissue protein levels in the recipient after transplantation, and/or focused on donor tissue after prolonged periods of warm and cold ischemia. None have focused on both a larger, more translationally relevant animal model and gene expression levels in the DBD before organ recovery. It is essential to better understand the state of potentially transplantable organs at the time of organ recovery, as this is when decisions are being made by transplant programs about the suitability of organs for transplantation. Combining these 2 approaches is necessary to inform potential human organ donor interventions aimed at increasing the availability of organs suitable for transplantation, as there may be targets for intervention identified in the donor.
Kidney transplantation is a common research target, since kidneys are the most frequently donated organs, and because obtaining biopsy data is a common practice. Further, delayed graft DGF, defined as a recipient needing hemodialysis within the 1st week after transplant, provides a short-term surrogate outcome for measuring graft success. What remains incompletely understood is how the cellular and molecular processes behind BD and its inflammatory response contribute to poor graft function and survival from DBDs. Previous research has shown that deceased organ donor kidney gene expression patterns are different than living donor specimens [Reference Hauser17], however, this work was performed after cold storage and was also limited by small sample size. The objective for our study was to better delineate the inflammatory response to herniation in the kidney using gene expression microarray technology to identify specific gene pathways that are altered before cold storage and transplantation. This study would be the first kidney-specific porcine model examining early changes in gene expression and gene pathways, and begins to provide correlative data for the inflammatory response to BD. Identification of specific inflammatory gene pathways could lead to future characterization of diagnostic biomarkers for organ quality, as well as the development of potential therapeutic interventions to improve graft function and survival. Based on their relevance to the cell-mediated immune response and transplantation, we hypothesized that cell adhesion molecule pathways involved in inflammation will be altered in the kidney following brainstem herniation.
Materials and Methods
Porcine Model of Brain Death
The animal protocols used in this study were approved by the UCI institutional animal care and use committee (UCI-IACUC). In total, 20 domestic female Chesterwhite or Yorkshire swine at 6 weeks of age were purchased from S&S Farms in San Diego County, CA. Animals (20–30 kg) were acclimated for 72 hours, fasted for 12 hours, and induced with an intramuscular combination of ketamine (21 mg/kg), xylazine (2.2 mg/kg), and atropine (0.04 mg/kg). Pentothal (10 mg/kg) was then given intravenously for induction via an ear vein catheter and endotracheal intubation was performed. Ventilation and anesthesia were maintained with isofluorane (0.75%–4%). Electrocardiogram leads, pulse oximetry, and end-tidal carbon dioxide (CO2) were monitored and adequate anesthesia was confirmed by assessing jaw tone, movement, and vital signs. Femoral artery and external jugular catheters were placed for hemodynamic monitoring, blood collection, and fluid administration. Based on a previously described model [Reference McLean18], a left-sided parietal burr hole was created, and a 15 cc epidural balloon catheter and subdural intracranial pressure (ICP) monitor were placed through this access point. A subdural laser Doppler flow probe was placed in the right frontal area (Fig. 1). After a 30-minute rest period, a physiologic baseline was established and brainstem herniation was induced by inflating the 15-cc balloon catheter over 1 min, mimicking the effects of a space-occupying lesion and eventual herniation. The balloon remained inflated for 20 minutes, then was deflated. Herniation was evaluated with a contralateral laser Doppler flow probe and an ipsilateral ICP catheter. BD was defined as a drop in blood flow to <15% of baseline, ICP persistently greater than mean arterial pressure (MAP), and fixed and dilated pupils [Reference McLean18]. BD was also verified in the first two animals in the BD group using conventional apnea testing in order to validate the previous model upon which our study was based [Reference McLean18]. Apnea testing was performed after preoxygenation with 100% oxygen for 5 minutes, followed by disconnecting the ventilator. Baseline levels of pCO2 were measured. After 10 minutes of being disconnected from the ventilator, another level of pCO2 was measured before placing the animal back on mechanical ventilation. If there was an increase of 20 mmHg pCO2 compared with baseline, BD was confirmed, consistent with national guidelines.
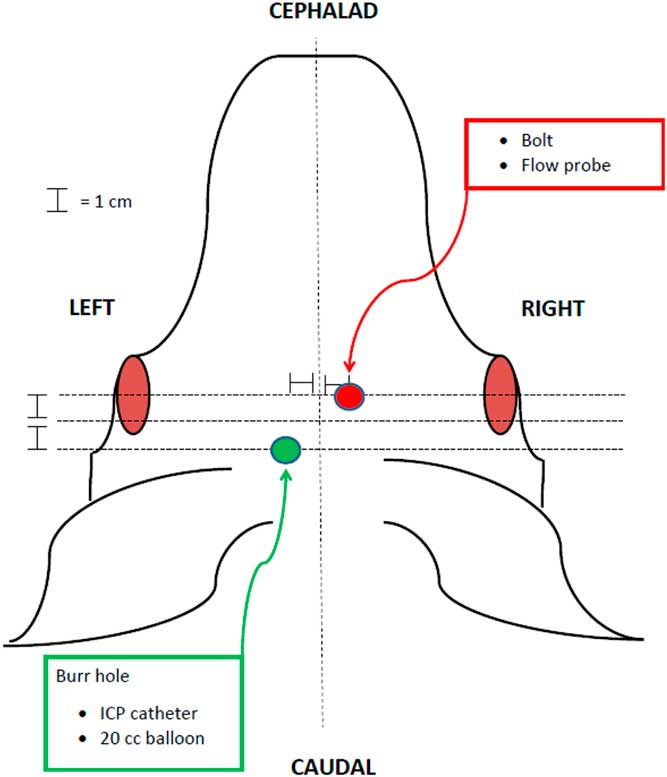
Fig. 1 Diagram of instrumentation of animals in the porcine herniation model. ICP, intracranial pressure
In total, 20 animals were randomized into 2 groups: a control group (burr hole, catheters, and anesthesia only) and a BD group (instrumentation plus inflation of subdural balloon catheter and confirmation of BD). Animals were supported on mechanical ventilation for 6 hours after randomization and 2 cc/kg boluses of 0.9% NaCl were given as needed to maintain a MAP above 35 mmHg to prevent cardiovascular collapse. Vasopressors were not utilized, consistent with the validated model upon which our study was based. Both groups were maintained with isofluorane for the duration of the case. A laparotomy was then performed and kidney specimens were obtained and snap frozen.
Vital signs were recorded every minute during the balloon inflation phase and at regular intervals during the 6-hour maintenance phase. Repeated measures ANOVA statistics were used to analyze mean physiologic variables and determine difference between groups during the balloon inflation phase and an independent-samples t-tests was used to analyze terminal values for ICP, MAP, and HR.
RNA Extraction
Total RNA for gene expression analysis was extracted from the porcine kidney tissue using TRIzol® (Gibco BRL Life Technologies, Rockville, MD, USA). RNA was purified using Qiagen-RNeasy Mini Kit. RNA pellets were resuspended in diethyl pyrocarbonate-treated water. RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Palo Alto, CA, USA). All samples had RNA Integrity Number (RIN) ≥9.1.
Gene Expression Microarrays
Microarray processing was performed as recommended by the manufacturer and is available in the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA, USA). In brief, first-strand cDNA was synthesized from 250 ng of total RNA. After making the complementary second strand, the double-stranded cDNA is used to generate biotin-tagged cRNA from an in-vitro transcription using T7 RNA polymerase. Ten μg of fragmented target cRNA was hybridized on an Affymetrix GeneChip® Porcine Genome Array. Arrays were scanned using GeneChip® Scanner 3000 7G and Command Console Software v 3.2.3. to produce .CEL intensity files.
Gene Expression Data Analysis
The results were analyzed using GeneSpring GX 12.1 Software (Agilent Technologies, Inc.). Raw data were normalized using GC-RMA. Only probe sets that reached a signal value ≥50 in at least 50% of the values in anyone out of the 2 conditions were included in the analysis. Overall, 17,566 of 54,675 probe sets represented on the array met these criteria. The microarray cell files and GC-RMA normalized data have been deposited in the GEO database (series accession number https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94709). Traditional Student’s paired t-test was first applied to each probe set and fold change (increase or decrease) >1.5 and Multiple Testing Correction (false discovery rate [FDR]<0.05) (Benjamini–Hochberg) procedure was carried out for statistical analysis.
The final list of significantly different expression probe sets between the 2 groups was then additionally analyzed using the functional annotation tools provided by DAVID 6.8, the Database for Annotation, Visualization and Integrated Discovery to classify the genes into pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Only pathways with Expression Analysis Systematic Explorer (EASE) score ≤0.05 are presented in this analysis. The EASE score is a modified Fisher exact p value in the DAVID system used for gene-enrichment analysis. An EASE score p value=0 represents perfect enrichment. p value ≤0.05 is considered as gene enrichment in a specific annotation category (http://david.abcc.ncifcrf.gov/helps/functional_annotation.html#summary).
Microarray Results Corroboration Using RT-PCR
For confirmation of gene expression microarray findings, reverse transcriptase polymerase-chain reaction (RT-PCR) assays were carried out on 6 genes from the KEGG cell adhesion pathway (SLA-DRA, SLA-1, SLA-DQA, ITGB2, ITGB8, and VCAM1). Analysis was performed with the Applied Biosystems 7900HT PCR System by using TaqMan Universal PCR Master Mix and Assays-on-Demand Gene Expression probes (Applied Biosystems) (SLA-DRA: assay ID, Ss03389945_m1; ITGB2: assay ID, Ss03392626_u1; ITGB8: assay ID, Ss03385280_g1; and SLA-1: Ss03395429_s1; SLA-DQA: Ss03389954_g1; VCAM1: assay ID, Ss03390914_m1). Actin β was used as an endogenous control.
Results
Animal Model
In total, 10 animals were assigned to the BD group and 10 animals were assigned to the control group. One animal in the BD group and 2 animals in the control group were unable to be maintained for 6 hours under general anesthesia and suffered cardiac arrest; upon autopsy, it was discovered that these animals had pre-existing illnesses.
Hemodynamic and intracranial physiologic results during the balloon inflation phase are displayed in Fig. 2. All animals in the experimental group and none in the control group met criteria for BD. Animals in BD and control groups had similar physiologic variables at initiation of general anesthesia: control animals had a mean heart rate (HR) of 80.3+18.4 beats per minute (BPM) and experimental animals had a mean HR of 85.1+9.0 BPM (p=0.49); MAP at induction was 64.6+11.6 mm of mercury (mmHg) for control animals and 68.7+11.0 mmHg for experimental animals (p=0.47); and ICP at induction was 17.3+5.5 mmHg for control animals and 16.8+7.3 mmHg for experimental animals (p=0.88).
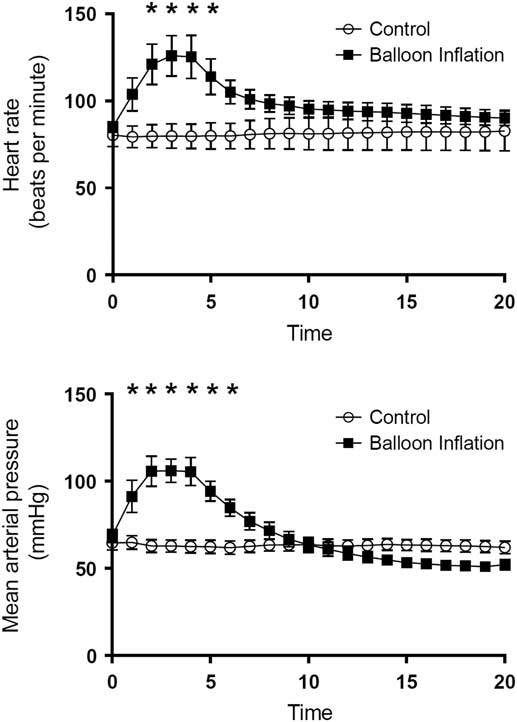
Fig. 2 Graphical representation of the physiologic response to inflation of balloon catheter (mimicking a herniation event) in animals. This graph shows time-dependent heart rate and mean arterial pressure measurements, stratified by control and brain death groups. *Statistically significant difference by ANOVA testing.
The expected catecholamine surge associated with herniation was demonstrated in the animals in the BD group during the time of balloon inflation. Initially, HR and MAP increased dramatically to a highest mean HR of 125.3±30.8 BPM at 4 minutes after balloon inflation and a highest MAP of 106.1±16.9 at 3 minutes postinflation. Means were compared using repeated measures ANOVA, and significant differences were found for MAP at minutes 2–6 and for HR at minutes 3–5 (Fig. 2) during balloon inflation are displayed in Fig. 2. At the conclusion of the balloon inflation phase, there were no significant differences between the groups with regard to HR or MAP (Fig. 2).
After maintenance of 6 hours of general anesthesia, the terminal ICP values were significantly different between control and experimental groups (19.3±4.5 vs. 59.3±18.8, p<0.001), however mean MAP (59.7±17.1 vs. 56.9±13.8, p=0.73) and HR (85±19.3 vs. 122.3±43.8, p=0.06) between groups were not significant.
The Effects of BD on Kidney Gene Expression
Using FDR<0.05 with 95% confidence, a total of 902 probe sets were differentially expressed between BD and control group which represent 233 annotated genes. In total, 139 genes had higher expression in the BD compared with control, and 94 genes had lower expression in the BD compared to control (online Supplementary Table 1).
We classified the final list of genes with significantly different expression between the 2 groups into pathways using the KEGG database. In total, 40 pathways (EASE score≤0.01, FDR<0.05) were enriched with genes that expressed significantly different between the 2 groups. Table 1 presents 11 selected pathways linked to the immune response, cell communication, allograft rejection, and graft-versus-host disease. Table 2 presents the individual upregulated genes within our primary pathway of interest, cell adhesion molecules, and the degree of fold change associated with the upregulation. Fig. 3 is a diagrammatic representation of the KEGG pathway for the cell adhesion molecules. In this diagram, the various genes and their interactions are displayed and those upregulated after BD in our animal model are noted.

Fig. 3 Diagrammatic representation of the interactions among genes within the cell adhesion molecule Kyoto Encyclopedia of Genes and Genomes pathway, with significantly upregulated genes in our experiments annotated. *Upregulated in porcine brain death model.
Table 1 Selected Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in porcine model significantly upregulated 6 hours following brainstem herniation when compared with control animals

EASE, expression analysis systematic explorer; FDR, false discovery rate; Jak-STAT, Janus kinase/signal transducers and activators of transcription; PPAR, peroxisome proliferator-activated receptor; UC, ulcerative colitis; SCI, spinal cord injury; TNF, tumor necrosis factor.
Table 2 Individual genes in Kyoto Encyclopedia of Genes and Genomes pathways significantly affected 6 hours following brain death when compared with sham group

FC, fold change; TNF, tumor necrosis factor.
RT-PCR was performed to corroborate the microarray results in our primary outcome of interest, the cell adhesion molecule pathway. A total of 6 representative genes were upregulated, 5 of which were noted to be significant (p<0.05) (Table 3).
Table 3 Reverse transcriptase polymerase-chain reaction verification of specific genes in the cell adhesion pathway

DISCUSSION
Efforts are needed to improve the quantity and quality of organs available for transplantation, and a better understanding of the inflammatory response to brainstem herniation may help to identify targets for future therapeutic interventions. Toward that end, we examined the genomic response to herniation in the porcine kidney and found significant differences in the expression levels of 233 genes, which were classified into gene pathways. These affected pathways play a central role in inflammation, allograft rejection, antigen processing and presentation, Toll-like receptor signaling, tumor necrosis factor (TNF) signaling, graft-versus-host disease, cell adhesion molecules, natural killer cell-mediated toxicity, Janus kinase/signal transducers and activators of transcription (Jak-STAT) signaling, peroxisome proliferator-activated receptor signaling, and chemokine signaling. Additionally, the results confirmed our hypothesis that adhesion molecule pathways are significantly upregulated after brainstem herniation and BD.
It has been established that a cascade of events result from severe neurologic injuries and brainstem herniation. This cascade usually begins with activation of the parasympathetic pathway, leading to hypothermia, hypotension, and electrolyte disturbances. This derangement is then followed by sympathetic stimulation, resulting in high catecholamine release, tachycardia, and hypertension [Reference Dziodzio, Biebl and Pratschke10]. Though this response to BD is well described in the literature, there is a dearth of information concerning the inflammatory response to brainstem herniation at the genetic level.
In reviewing the genetic contribution to organ transplantation after BD, both existing genotypes and regulation of these genotypes, or epigenetic events, should be examined. Several groups have examined the idea that certain genetic makeups are predictive of success in transplantation. One early study, limited by sample size, was unable to show any association between donor single-nucleotide polymorphisms and DGF [Reference Israni28]. However, subsequent studies have shown that differences in TNF-α and TGF-β gene polymorphisms between donor and recipient were associated with increased acute rejection [Reference Alakulppi29, Reference Park30]. The fact that genetic polymorphisms are associated with differential rejection rates suggests that examining downstream gene expression patterns may increase our understanding of BD and how it affects transplantation outcomes.
In addition to polymorphisms that may contribute to graft function, gene pathways that are affected during the process of BD have been described in the literature. However, these previous genomic studies have been performed on tissue immediately prior to implantation, after prolonged periods of both warm and cold ischemia. In our model, we examine the effects of BD on the genomic pathways in the donor, prior to any period of ischemia, cold storage, or interaction with the recipient immune response. With respect to the pre-implantation literature, several groups have examined apoptotic pathway, and DGF has been shown to correlate with increased expression in biopsies obtained after cold storage [Reference Goncalves-Primo31]. Confounding these results is that fact that longer durations of CIT have been shown to increase apoptosis expression in specimens from DBDs [Reference lznerowicz7]. In another study, higher pre-implant lipocalin 2 (LCN-2) expression correlated with increased DGF rates and acute rejection episodes [Reference Kaminska32]. In an effort to avoid alterations in gene expression patterns that cold ischemia may induce, our animal model focuses on tissue specimens obtained at the time of organ recovery, prior to cold preservation, to isolate those pathways strictly related to the process of brainstem herniation that could be targets for intervention during organ donor management.
Many of the described studies have been performed in mice or rats, which have poor genetic correlation with human pathways [Reference Seok33]. Due to both greater genetic and anatomic similarities with humans, swine are one of the most commonly used species in biomedical translational research models. In 2012, Hume et al. published the first porcine genome-wide transcriptional analysis that included 62 tissues and cell types, providing an important resource for understanding the relationship between porcine and human gene expression [Reference Freeman34]. In one of the first porcine models of brainstem herniation, Mclean et al. examined the effect of glucocorticoid administration on circulating levels of pro- and anti-inflammatory cytokines, and found that glucocorticoids shifted toward a more anti-inflammatory state [Reference McLean18]. Additional studies have examined mRNA and protein levels in heart, liver, and kidney tissues, looking at TNF-α, IL-6, and IL-10 in one study [Reference Barklin35] and TNF-α, IL-6, IL-1β, IL-6R, ICAM-1, MCP-1, and TGF-β in another [Reference Skrabal36]. These studies have helped to delineate the inflammatory response to BD, and may explain organ-specific differences in transplantation outcomes after BD. However, variable results were obtained, with the former study did not show increased levels of cytokine mRNA between organ tissues [Reference Barklin35], whereas the latter study showed a mixed picture of cytokine upregulation in the studied organs [Reference Skrabal36]. Whereas these studies have addressed targeted genes, we have sought to take a more global approach and use microarrays to identify a large numbers of genes simultaneously, identify their molecular interaction and classify them to biological gene pathways. Perhaps most similar to our study is a comparison between BD and living donor swine that examined differences in apoptotic and protective gene expression patterns in multiple tissues, albeit after cold storage [Reference Stiegler37].
In our porcine model of BD, we found that 233 unique genes were noted to be significantly altered in those animals subject to brainstem herniation, when compared with those in a control group. These genes were classified into KEGG gene pathways that are related to immune modulation. The immune system is responsible for the acute [Reference Pratschke13] and chronic [Reference Pratschke38] rejection of organs, and cell adhesion molecule upregulation was hypothesized to be particularly affected. Cell adhesion molecules (CAMs) are glycoproteins expressed on the cell surface and play a critical role in a wide array of biologic processes that include hemostasis, the immune response, inflammation, embryogenesis, and development of neuronal tissue. They have been shown to be integral in ischemia-reperfusion (I:R) injuries in kidney tissue [Reference Kusaka39], and increased levels of CAMs have been associated with increased mortality in transplant recipients [Reference Connolly40]. Several publications have examined treating donors to mitigate the I:R injury, recently proposing that CAM-mediated I:R injury can be treated with N-octanoyl dopamine. In this study, the researchers showed better early graft function and reduced acute rejection in renal allograft recipients after donors were treated with the drug [Reference Spindler41]. These results were obtained in rats, and further investigation is warranted.
Ten additional gene pathways related to the immune system were significantly affected by the process of herniation in our study. Antigen processing and presentation has been associated with macrophage presence in rejected human renal allograft specimens [Reference Bergler19], and antibody-mediated rejection has been a target of anti-rejection medications for years. Toll-like receptor-4 (TLR-4) has been increasingly recognized as playing a critical role in the pathogenesis of I:R injury of renal grafts [Reference Krichen42, Reference Zhao43], and research into anti-TNF agents have shown promise with regards to being protective against I:R injury in animal models [Reference Nagata21]. Additionally, we identified pathways associated with common immunologic processes affecting renal transplantation and rejection, such as graft-versus-host disease [Reference Chen44], natural killer cell-mediated toxicity [Reference Kohei45], Jak-STAT signaling [Reference Baan46], peroxisome proliferator-activated receptor signaling [Reference Kiss47], chemokine signaling, and allograft rejection.
The limitations of this study include correlative data only. Though unable to prove causation, these data are a first step in identifying gene pathways that are altered in kidney tissue after BD. Additionally, there are no internal controls performed prior to herniation in each specimen, which potentially would aid in the delineation of the inflammatory response. Isofluorane anesthesia also may be considered to alter the inflammatory response. However, we attempted to control for this by administering this anesthetic to both the experimental and control animals, even though it was not needed for sedation or comfort in the BD group. It is also unclear as to the ultimate source of the inflammatory response to BD. Some investigators have postulated that neutrophils, platelets [Reference Stahel48], and the nervous system [Reference Fluiter49] may be sources of the inflammatory response, but we were unable to address these concepts in the current study. Our intent was to address only the kidney, and future work would involve analysis of all transplantable organs, protein levels within organs, and circulating markers. In addition, expanding the animal model to include allotransplantation of organs into another subject would allow correlations between donor gene expression/biomarker patterns and recipient graft outcomes, the ultimate consequence of interest. This expansion would facilitate investigation of whether interventions in the donor can abrogate the inflammatory response and improve transplantation outcomes.
In this study, we addressed only the 6-hour time point after herniation. This time point was chosen for this initial study for several reasons. This time point is a practical surrogate for when actual interventions could begin to be carried out in a donor. In general, declaration of BD occurs 1–2 hours after the actual BD event, and then it requires several additional hours of obtaining authorization for donation and examining the donor before the organ procurement organization can begin to make interventions. A 6-hour time point marks the approximate start of donor management by the organ procurement organization, when interventions in the donor possibly could begin. Future studies would examine later time points, further characterizing the gene expression and gene pathways affected by BD and potentially guiding therapeutic interventions at these times. The raw data of the 6-hour time point can be found in GEO database (series accession number https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94709) and is available for additional analyses and comparisons with current and future genomic datasets related to kidney injury or other kidney donor studies. Many studies have shown conflicting results in examination of the impact of duration of donor management [Reference lznerowicz7, Reference Muruve50–Reference Nijboer52] and the best strategy for optimal duration of management is still under investigation. Later time points in a porcine model may help in determining kidney-specific genomic changes in examining duration of donor management.
Using a porcine model of brainstem herniation, we showed for the first time that multiple gene pathways associated with inflammation and organ rejection are altered in the donor after BD when compared with control animals. Building on this approach, new studies will generate data on additional time points with a goal to characterize potential biomarkers and gene pathways and identify the window of opportunity for organ management/procurement and. Animal models that correlate gene expression patterns in organs at the time of recovery with post-transplant outcomes would be essential in identifying appropriate targets in the DBD for future interventions. With the development of valid animal models, possible interventions could be tested with little risk to transplantable human organs. Donor management after BD is one step in the process of organ donation in which these interventions could occur. This study begins to identify the underlying molecular mechanisms associated with this event, and aims to act as a first step towards targeting specific pathways that could optimize organs for successful transplantation.
Supplementary materials
To view supplementary material for this article, please visit https://doi.org/10.1017/cts.2018.312
Acknowledgements
This paper was partially supported by NIH-NCATS grant no. UL1 TR001414.
Disclosures
The authors have no conflicts of interest to declare. The contents of this article do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.








