Introduction
Cystic Echinococcosis (CE) is a neglected zoonotic disease caused by Echinococcus granulosus sensu lato (McManus Reference McManus, Eckert J, Meslin and Pawlowski2001). CE is a highly prevalent parasitic disease worldwide, particularly in regions where people are involved in animal farming and come into contact with free-roaming or herding dogs (McManus Reference McManus, Eckert J, Meslin and Pawlowski2001). The World Health Organization has incorporated CE into its strategic plan for neglected diseases, to prevent, control, eliminate, and eradicate these diseases by 2030 (Worl Health Organization 2020). Infection typically starts silently, and the clinical presentation of individuals with CE can vary from being entirely asymptomatic to experiencing severe illness. While hydatid cysts can potentially develop in any organ, approximately 80% of the patients have a single cyst either in the liver (4/5) or in the lungs (1/5) (Brunetti et al. Reference Brunetti, Kern and Vuitton2010; Brunetti et al. Reference Brunetti, Tamarozzi, Macpherson, Filice, Piontek, Kabaalioglu, Dong, Atkinson, Richter, Schreiber-Dietrich and Dietrich2018). Most cases go unnoticed for extended periods and are often diagnosed incidentally (Eckert et al. Reference Eckert, Gemmell, Meslin, Pawlowski and World Health2001; Tamarozzi et al. Reference Tamarozzi, Covini, Mariconti, Narra, Tinelli, De Silvestri, Manzoni, Casulli, Ito, Neumayr and Brunetti2016). Diagnosis and treatment in humans do not impact parasite transmission, but they are crucial in increasing awareness and mitigating the disease’s overall burden (Tamarozzi et al. Reference Tamarozzi, Deplazes and Casulli2020).
CE diagnosis still relies on imaging techniques (mainly US), with MRI and CT being applied when US is not feasible or when further clinical information is needed. The World Health Organization’s Informal Working Group on Echinococcosis (WHO-IWGE) has established a globally recognized classification system for CE cysts based on the sonographic appearance of the cysts. This classification categorizes CE cysts into six distinct stages, which are further grouped into three clinical categories: active (CE1 and CE2), transitional (CE3a and CE3b), and inactive (CE4 and CE5) (Enrico Brunetti et al. Reference Brunetti, Tamarozzi, Macpherson, Filice, Piontek, Kabaalioglu, Dong, Atkinson, Richter, Schreiber-Dietrich and Dietrich2018). Although CE3a and CE3b are classified as ‘transitional’ stages, in practice, CE1, CE2, and CE3b are unequivocally considered active cysts, with no doubt regarding their vitality. Over time, it has been determined that approximately 50% of CE3a cysts are indeed alive (Hosch et al. Reference Hosch, Junghanss, Stojkovic, Brunetti, Heye, Kauffmann and Hull2008). Hence, distinguishing between the active category (CE1, CE2, CE3a, and CE3b) and cysts that are presumed to be mostly no longer viable (CE4 and CE5) is of utmost importance. This classification forms the cornerstone for tailoring treatment approaches according to the specific stage of the cyst. Treatment options for CE include a range of modalities, including surgical procedures, percutaneous interventions such as PAIR, standard catheterization, and modified catheterization technique (MoCaT), drug therapies, and ‘watch and wait’ approach (Akhan Reference Akhan2023; Akhan et al. Reference Akhan, Ozmen, Dinçer, Sayek and Göçmen1996; Enrico Brunetti et al. Reference Brunetti, Tamarozzi, Macpherson, Filice, Piontek, Kabaalioglu, Dong, Atkinson, Richter, Schreiber-Dietrich and Dietrich2018; Kern et al. Reference Kern, Menezes da Silva, Akhan, Müllhaupt, Vizcaychipi, Budke and Vuitton2017). Serological tests are mostly employed when the imaging results are not pathognomonic (Tamarozzi et al. Reference Tamarozzi, Covini, Mariconti, Narra, Tinelli, De Silvestri, Manzoni, Casulli, Ito, Neumayr and Brunetti2016). The sensitivity and specificity of serological tests for CE disease are known to vary based on numerous factors. Studies have reported that the sensitivity of these tests can vary influenced by factors such as patient characteristics and cyst characteristics, including location, number, and stage. False-negative test results may occur in hepatic CE cases, particularly with young CE1 cysts (30–58%), inactive CE4-CE5 cysts (50–87%), and in cases of extra-hepatic CE, including up to 50% of patients with lung cysts and cysts in other locations (Hernández-González et al. Reference Hernández-González, Santivañez, García, Rodríguez, Muñoz, Ramos, Orduña and Siles-Lucas2012; Lissandrin et al. Reference Lissandrin, Tamarozzi, Piccoli, Tinelli, De Silvestri, Mariconti, Meroni, Genco and Brunetti2016; Ortona et al. Reference Ortona, Riganò, Margutti, Notargiacomo, Ioppolo, Vaccari, Barca, Buttari, Profumo, Teggi and Siracusano2000; Siles-Lucas et al. Reference Siles-Lucas, Uchiumi and Tamarozzi2023). Patients with transitional (CE3a, CE3b) and CE2 cysts exhibit lower seronegativity rates (5–20%), while those with multiple cysts are typically seropositive (Lissandrin et al. Reference Lissandrin, Tamarozzi, Piccoli, Tinelli, De Silvestri, Mariconti, Meroni, Genco and Brunetti2016).
Circulating cell-free DNA (cfDNA) is being explored as a minimally invasive biomarker (Khier and Lohan Reference Khier and Lohan2018). The utilization of cfDNA analysis has gained significant interest in clinical practice, especially in non-invasive prenatal testing, cancer detection, and follow-up, as well as in pathogen identification (Benn and Cuckle Reference Benn and Cuckle2023; Blauwkamp et al. Reference Blauwkamp, Thair, Rosen, Blair, Lindner, Vilfan, Kawli, Christians, Venkatasubrahmanyam, Wall, Cheung, Rogers, Meshulam-Simon, Huijse, Balakrishnan, Quinn, Hollemon, Hong, Vaughn, Kertesz, Bercovici, Wilber and Yang2019; Islam et al. Reference Islam, Gopalan, Lam and Shiddiky2023). The term cfDNA primarily encompasses nuclear and/or mitochondrial DNA, which constitutes a diverse mixture released from both healthy and unhealthy cells into body fluids such as blood (Petit et al. Reference Petit, Carroll, Gould, Pockney, Dun and Scott2019), saliva (Brooks et al. Reference Brooks, Malkin, De Michino and Bratman2023), urine (Kueng et al. Reference Kueng, Arcioni, Sandberg, Kuhn, Banz, Largiadèr, Sidler and Amstutz2023), cerebrospinal fluid, etc. (Afflerbach et al. Reference Afflerbach, Rohrandt, Brändl, Sönksen, Hench, Frank, Börnigen, Alawi, Mynarek, Winkler, Ricklefs, Synowitz, Dührsen, Rutkowski, Wefers, Müller, Schoof and Schüller2023; Ozturk and Caner Reference Ozturk and Caner2022). The presence of parasite-derived cfDNA in bodily fluids is potentially promising in diagnosing various diseases, including malaria (Buppan et al. Reference Buppan, Putaporntip, Pattanawong, Seethamchai and Jongwutiwes2010), trypanosomiasis (Madrigal et al. Reference Madrigal, Marcus, Gilman, Scott and Shiff2019), leishmaniasis (de Almeida et al. Reference de Almeida, Koru, Steurer, Herwaldt and da Silva2017), schistosomiasis (Kato-Hayashi et al. Reference Kato-Hayashi, Yasuda, Yuasa, Isaka, Haruki, Ohmae, Osada, Kanazawa and Chigusa2013), strongyloidiasis (Javanian et al. Reference Javanian, Gorgani-Firouzjaee and Kalantrai2019), and echinococcosis (Moradi et al. Reference Moradi, Meamar, Akhlaghi, Roozbehani and Razmjou2019). As reviewed by Zhao et al., numerous studies have been published on the identification of Echinococcus-derived cfDNA in samples from individuals with echinococcosis (ZhaoShen et al. Reference Zhao, Gongsang, Ji, Li, Qi, Li, Qiangba, Danzeng, Chen, Zhou, Huasang, Yin, Pei, Xie, Cai, Asan, Pang, Li, Chen and Li2021). Nonetheless, these investigations have yet to definitively establish whether cfDNA could serve as a viable alternative to or prove more advantageous than serological tests. Additionally, it remains unclear whether cfDNA results align consistently with imaging findings in the diagnosis of CE. The primary aim of this study is to determine whether cfDNA targets (Primers for Echinococcus specific repeat sequences: mgs-4, mgs-12 and fragment located within a repetitive sequence: EG1 Hae III) have the potential to distinguish CE patients from healthy individuals and to evaluate the comparative results with a commercial E. granulosus-specific IgG enzyme-linked immunosorbent assay (ELISA), which is frequently used in routine diagnosis (Tamarozzi et al. Reference Tamarozzi, Silva, Fittipaldo, Buonfrate, Gottstein and Siles-Lucas2021). Additionally, the study assessed its potential for distinguishing between active and inactive CE patients.
Materials and Methods
Study group and sample collection
The study was designed as a prospective study, and 76 hepatic CE patients (43 female and 33 male) were included in the study after the abdominal US examination. Radiological findings were accepted as the gold standard when determining the patient group. Hence, the radiological data were reviewed by two independent radiologists with a minimum of 5 years of experience in the diagnosis, treatment, and follow-up of CE. The radiologists evaluated independently without knowledge of each other’s findings. A third radiologist’s opinion was asked if further analysis or consensus was necessary. The cases were classified as active (CE1, CE2, CE3a, CEb) or inactive (CE4, CE5) CE according to the WHO-IWGE classification. Patients with hydatid cysts localized in organs different than the liver, those with multiple hydatid cysts of different groups (active and inactive), and those who had previously received any treatment for CE (surgical, percutaneous, or medical) were excluded from the study. Transitional cysts have been considered active in this study due to their indication for treatment. In addition, 43 healthy individuals (24 female and 19 male, not being diagnosed with any metabolic, autoimmune, cardiovascular, malignancy, or infectious disease) were included in this study. Blood samples were collected from healthy individuals and patients at the initial diagnosis, and serum was obtained and stored at -80°C until DNA extraction.
DNA extraction and cfDNA detection
Total DNA extraction was carried out from the serum (starting volume 200 μl) of the participants using Exgene Clinic SV (GeneAll) kit following the manufacturer’s instructions. The concentration and purity of the DNA were measured by FLUOstar Omega Microplate Reader (BMG LABTECH) using LVis plate. Real Time-PCR was performed using SYBR Green Master Mix (ABT) and primers at 200 nM final concentration (Primers for Echinococcus specific repeat sequences: mgs-4 (~83 bp) and mgs-12 (~87 bp)) with ViiA™ 7 Real-Time PCR System (Thermo Fisher Scientific) (Wan et al. Reference Wan, Peng, Ma, Tian, Wu, Li, Ling, Lv, Ding, Tan and Zhang2020). The cycling protocol was as follows: initial denaturation for 10 minutes at 95°C, followed by 45 cycles of denaturation at 95°C for 10 seconds, and annealing at 60°C for 30 seconds. A nested PCR was conducted to specifically amplify a 133 bp fragment located within a repetitive sequence (EgG1 Hae III) present in the genome of E. granulosus s.l (Abbasi et al. Reference Abbasi, Branzburg, Campos-Ponce, Abdel Hafez, Raoul, Craig and Hamburger2003). The PCR process involved two rounds of amplification, and the following conditions were employed: In the first round of amplification, the primers Eg2691 and Eg2692 were utilized to target the repetitive sequence, resulting in the generation of a 269 bp PCR product. This product served as the template for the second round of amplification. In the second round of amplification, a different set of primers, Eg1121aF and Eg1122aR, were employed to specifically amplify the diagnostic band of interest, which measures 133 base pairs (Al-Hindi et al. Reference Al-Hindi, Bodell and Alshmmari2023). DNA amplification was carried out using a thermal cycler under the following conditions: an initial denaturation step at 95°C for 5 minutes, followed by 35 cycles of 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute, concluding with a final elongation step at 72°C for 10 minutes (Boufana et al. Reference Boufana, Campos-Ponce, Naidich, Buishi, Lahmar, Zeyhle, Jenkins, Combes, Wen, Xiao, Nakao, Ito, Qiu and Craig2008). Sequences of the primer sets used in the study are presented in Table 1. The selected amplicons for each target were confirmed by Sanger sequencing.
Table 1. Details of the primer sets

Serological tests
Obtained sera were evaluated using the commercially available serological test Hydatidosis IgG ELISA (Vircell SL, Granada, Spain) according to the manufacturer’s instruction. All results were reported as positive or negative. All tests were performed in the same session. Borderline results were accepted as negative to reduce the risk of false-positive diagnoses.
Statistical analyses
The study was designed with a power of 90% and a Type I error rate of 5%, based on an anticipated effect size of 0.3, to determine the required sample size for each group (active cyst, inactive cyst, and healthy control). Statistical analysis and calculations were performed using IBM SPSS Statistics Version 23.0 (IBM Corp., NA, USA) program. The normal distribution of the variables was analyzed graphically and by the Shapiro-Wilk test. Descriptive data were presented as either mean ± standard deviation, frequencies, or percentages as appropriate. One-way ANOVA was used to compare continuous variables when parametric test assumptions were met. The chi-square test or Fisher exact test was used to compare categorical variables. US classification of cysts was considered as gold standard, and performance measures (sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy) of other tests were calculated according to the gold standard. The performance of the tests was compared by using the McNemar test. In statistical decisions, p≤0.05 was accepted as an indicator of significant difference. Bonferroni correction was applied for multiple comparisons.
Results
Characteristics of patients
Patients were allocated to groups based on the WHO-IWGE classification system. The number of the patients in active and inactive cyst groups was 34 and 42, respectively. The majority of the patients were female (43/76, 56.6% respectively). The patients’ ages ranged from 18 to 82 years, with a mean age of 42 years. The majority of the patients (active CE and inactive CE) had a single cyst (23/34, 71.8% and 26/42, 61.9%). For patients with multiple CE cysts, all cysts were either all active or all inactive. The mean size of cysts was 6.89 cm. Cyst size (≥10, giant cyst) was recorded to be significantly higher in patients with active CE (Chi square test, p<0.001). Table 2 presents all the characteristics of the patients.
Table 2. The characteristics of the patients

cfDNA detection
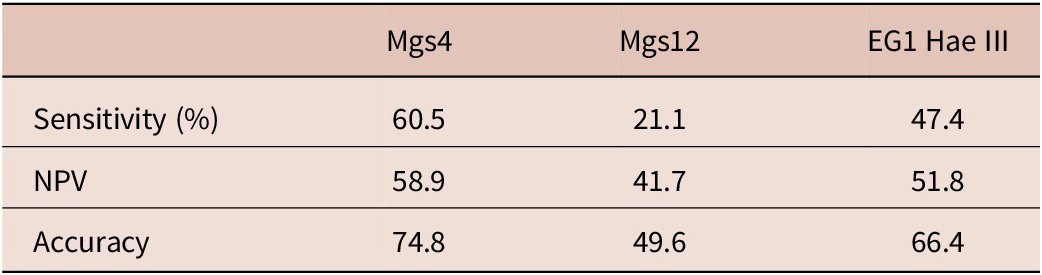
Results from the analysis of the cfDNA-targeted tests revealed that mgs-4 was the most effective diagnostic target in distinguishing between CE patients and healthy controls, with a sensitivity of 60.5% (McNemar test, p = 0.003). Following, EG1 Hae III demonstrated a sensitivity of 47.4%, while mgs-12 had a sensitivity of 21.1%. The specificity and PPV of all cfDNA targeted tests was found as 100%. The sensitivity, NPV, and accuracy values of the cfDNA-targeted tests are given in Table 3.
Table 3. The performance measures of the cfDNA-targeted tests for control group and CE patients
In the comparative analysis of active and inactive patient groups against healthy controls concerning cfDNA targets, 100% specificity and PPV were discerned in both patient categories. However, the sensitivity of these cfDNA targets displayed variability among groups. Among active CE patients, sensitivity rates were 52.9%, 23.5%, and 52.9% for the mgs-4, mgs-12, and EG1 Hae III assays, respectively. In contrast, patients with inactive cysts exhibited sensitivity rates of 66.7%, 19%, and 42.9% for the same assays. The sensitivity, NPV, and accuracy values of the cfDNA-targeted tests obtained as a result of the comparative analysis of the active and inactive CE patient groups and the control group are given in Table 4. The real-time PCR results for mgs-4 and mgs-12 are given in Figure 1.
Table 4. The performance measures of the cfDNA-targeted tests of the active and inactive CE patients compared to the control group


Figure 1. Real-time PCR results for mgs-4 and mgs-12 showing overall positivity and negativity rates across control, active CE, and inactive CE groups.
Serodiagnosis
The ELISA yielded negative results for all participants in the control group, while it showed positive results in 18 of the 34 (52.9%) active and 9 of the 42 (21.4%) inactive CE patients. The ELISA demonstrated a specificity of 100% and a sensitivity of 35.5% when distinguishing between patients with CE and healthy controls. The PPV, NPV, and accuracy values of the ELISA were found to be 100%, 46.7%, and 58.8%, respectively. The seropositivity rate was significantly higher in the active compared to inactive CE groups (McNemar test, p=0.024). There was no relationship between ELISA results and cyst features such as cyst number, diameter, and location (Chi-square test, p> 0.05).
Combination of multiple diagnostics
Combining ELISA with the mgs-4 target led to an increased sensitivity of 72.4% and improved accuracy of 82.4% for distinguishing between CE patients and the control group. Table 5 presents the sensitivity, NPV, and accuracy values obtained through the combination of ELISA with various other cfDNA targets. Specificity and PPV were found to be 100% for all combinations.
Table 5. The performance measures of the combination of ELISA with various other cfDNA-targeted tests for distinguishing between CE patients and the control group

In the assessment of test combinations within active CE patients and healthy controls, it was observed that the combination of ‘ELISA+mgs-4’ or ‘ELISA+ EgG1 Hae III’ elevated sensitivity to 70.6% and accuracy to 87%. When discerning between inactive CE patients and healthy controls, the combined utilization of ELISA and the mgs-4 test resulted in an increased sensitivity of 73.8% accuracy to 87.1%. Figure 2 presents the sensitivity values derived from the combination of ELISA with different cfDNA targets.

Figure 2. Sensitivity values for all tests and combinations in study groups.
Discussion
The study aimed to assess the potential of Echinococcus cfDNA as an adjunct diagnostic test for CE in comparison to serology. The evaluation of the tests’ diagnostic potential involved utilizing various combinations of the tests used. To accomplish this objective, three specific targets for cfDNA detection – namely, partial EgG1 Hae III, mgs-4 and mgs-12 – were evaluated. These primer pairs were chosen to improve template utilization efficiency and restrict the length of resulting products to fall within the range of 83 to 133 base pairs. This constraint considers the typical size of cell-free DNA (cfDNA), which is approximately 166 base pairs (Wan et al. Reference Wan, Peng, Ma, Tian, Wu, Li, Ling, Lv, Ding, Tan and Zhang2020).
Accurate diagnosis plays a pivotal role in the effective management of CE, with the identification of stage-specific treatment strategies essential to optimize outcomes for the highest benefit of patients. Currently, imaging techniques, particularly US, stand as the predominant and dependable method for diagnosing CE. Serological tests play a supporting role in the diagnostic process, but their results can vary and are not as definitive as imaging modalities (Siles-Lucas et al. Reference Siles-Lucas, Uchiumi and Tamarozzi2023; Tamarozzi et al. Reference Tamarozzi, Akhan, Cretu, Vutova, Akinci, Chipeva, Ciftci, Constantin, Fabiani, Golemanov, Janta, Mihailescu, Muhtarov, Orsten, Petrutescu, Pezzotti, Popa, Popa, Popa, Velev, Siles-Lucas, Brunetti and Casulli2018). The importance of conducting tests to confirm imaging findings is unquestionable. In the studies examining the relationship between serology and clinical features, the most evaluated variable is the cyst stage. Many studies assessing the accuracy of serological tests have primarily focused on determining the presence or absence of infection, often without considering other relevant clinical variables. However, a consistent finding across various studies is that patients with CE1 and CE4-CE5 (hepatic) cysts frequently test seronegative, with percentages ranging from 30% to 58% and 50% to 87%, respectively. In contrast, the rates of seronegativity are lower when CE2 and CE3 (CE3a-CE3b) cysts are present, ranging from 5% to 20% (Brunetti et al. Reference Brunetti, Tamarozzi, Macpherson, Filice, Piontek, Kabaalioglu, Dong, Atkinson, Richter, Schreiber-Dietrich and Dietrich2018; Lissandrin et al. Reference Lissandrin, Tamarozzi, Piccoli, Tinelli, De Silvestri, Mariconti, Meroni, Genco and Brunetti2016; Tamarozzi et al. Reference Tamarozzi, Silva, Fittipaldo, Buonfrate, Gottstein and Siles-Lucas2021). This study determined that the serological sensitivity in distinguishing CE patients from healthy controls was quite low at 35.5%. However, when focusing solely on patients with active CE cysts, the sensitivity of the ELISA completely in line with the literature increased to 52.9%. In contrast, the sensitivity was lower (21.4%) for detecting inactive CE when compared to healthy controls. These findings are consistent with previous research and highlight the need for supplementary diagnostic tests, especially for patients with inactive cysts.
In rural areas where healthcare accessibility is scarce and the prevalence of certain diseases like CE is high, the adoption of current diagnostic tools like cfDNA offers tremendous potential to improve the precision and swiftness of disease detection (ZhaoShen et al. Reference Zhao, Shen, Jin, Wang, Li and Chen2021). Because of its capacity to achieve a highly accurate early diagnosis, cfDNA is currently being extensively investigated and has been identified in the detection of various parasitic infections, including CE (Weerakoon and McManus Reference Weerakoon and McManus2016). Due to the increasing focus on cfDNA-based methodologies, numerous investigations have been undertaken to ascertain the presence and quantify Echinococcus cfDNA within the host’s serum/plasma and urine (ZhaoShen et al. Reference Zhao, Shen, Jin, Wang, Li and Chen2021). In the first study published on CE, cfDNA targeting the NAD1 region (450 pb) of E. granulosus was examined in a cohort of 25 CE patients and 25 individuals as the control. The findings revealed that 20% (5/25) of serum samples from the CE patients with ruptured cysts resulted positive for cfDNA, while no positivity was observed in either the urine samples or the control group (Chaya and Parija Reference Chaya and Parija2014). In contrast to the findings of this study, we detected cfDNA positivity in patients with non-ruptured CE cysts as well. This discrepancy may be attributed to differences in target nucleic acid size, which can influence sensitivity. Our results suggest that cfDNA detection may not be solely dependent on cyst rupture, highlighting the potential for broader diagnostic utility. In another study, cox1 (400 bp) and NAD1 (450 bp) regions were targeted in the parasite DNA derived from serum and paraffin-embedded tissue samples obtained from 80 CE patients, and at least one gene positivity was detected in only 19 serum samples. However, they concluded that their method of targeting longer fragments may have missed the mark because Echinococcus likely releases shorter DNA fragments (90–200 bp) into the bloodstream (Moradi et al. Reference Moradi, Meamar, Akhlaghi, Roozbehani and Razmjou2019). In the investigation of cfDNA (133 bp) in the urine of 12 CE patients and 25 healthy individuals, it revealed positive findings in 9 out of 12 patients, yielding a reported sensitivity of 75% (Toribio et al. Reference Toribio, Santivanez, Scott, Enriquez, Sedano, Soto-Becerra, Garcia and Shiff2020). Several recently published studies have used next-generation sequencing approaches to identify the structure and nature of parasite-derived cfDNA in the blood of patients for various echinococcosis clinics depending on different causative agents. It has been shown that nuclear and mitochondrial genome fragments of ~100 bp to 350 bp of circulating E.granulosus cfDNA in 22 CE patients could be mapped (Fan et al. Reference Fan, Gai, Zhang, Ma, Wang, Chen, Dong, Zhang, Bao, Zhou, Ren, Cairang, Hou, Ren, Wang, Wang and Song2021; Ji et al. Reference Ji, Li, Li, Danzeng, Li, Zhao, Qiangba, Zhang, Renzhen, Basang, Jia, Gongsang, Ma, Wang, Chen, Zhou, Huasang, Yin, Xie, Pei, Cai, Jiang, Yang, Wang, Asan, Han, Li, Chen and Yang2020; ZhaoGongsang et al. Reference Zhao, Shen, Jin, Wang, Li and Chen2021).
According to the obtained results, the best diagnostic cfDNA target was identified as mgs-4 with 52.9% and 66.7% sensitivity in patients with active and inactive cysts, as compared to the healthy control group, respectively. The exact reason for this higher sensitivity rate in inactive cysts is unclear; it may be due to the cysts releasing more cfDNA into the bloodstream as they die. The combination of mgs-4 and ELISA showed promise as a diagnostic reference for CE. For instance, the sensitivity results for ELISA, mgs-4, and the combined ELISA and mgs-4 in distinguishing between patients and healthy controls were 35.5%, 52.9%, and 72.4%, respectively. Furthermore, the sensitivity results for ELISA, mgs-4, and ‘ELISA+ mgs-4’ patients with active cysts, compared to the healthy control group, were established as follows: 52.9%, 52.9%, and 70.6%, respectively. In addition, the sensitivity for the identical targets in inactive CE patients, when compared to the healthy control group, was determined as follows: 21.4%, 66.7%, and 73.8%, respectively.
It is important to interpret the results of this study in the context of its limitations. First, we excluded patients with other parasitic diseases from our research. As the primary aim of this study was to determine whether the newly tested molecular targets could effectively distinguish between healthy individuals and CE patients, it was deemed essential to use healthy controls for this initial phase. Once the diagnostic value of these targets is established, additional control groups, including patients with other parasitic diseases or consider cysts in different organs, such as the lungs or kidneys, to establish the specificity of these findings. Another limitation of this study is that the EG1 Hae III primers were designed to target the G1 genotype of Echinococcus granulosus s.s., potentially limiting their ability to detect other genotypes within E. granulosus s.l‥Therefore, it is possible that the weak or negative results obtained could be due to patients being infected with a different genotype of E. granulosus, which was not detected by these primers. Further genotyping studies are needed to confirm this.
In conclusion, this study has successfully detected Echinococcus cfDNA circulating in the blood of individuals with CE. The mgs-4 target has exhibited superior sensitivity compared to commonly used commercially available serological test. When combined with ELISA, mgs-4 has shown significantly higher sensitivity for diagnosing CE across all group comparisons. While these tests may not completely replace serological tests, they hold substantial diagnostic value, especially in accurately diagnosing inactive patients. Molecular analyses could be implemented in clinical practice alongside serology, particularly in cases where serological results are inconclusive. Exploring the inclusion of additional molecular targets could further improve diagnostic accuracy and should be a focus for future research. This is crucial not only for differential diagnosis but also for collecting precise epidemiological data. Therefore, further testing of both these cfDNA targets and potential new ones in larger sample groups is necessary before their integration into the existing routine laboratory diagnostic protocol.
Data availability
Data available on request from the authors.
Financial support
This study was supported by Scientific and Technological Research Council of Turkey (TUBITAK) under the Grant Number 122S804. The authors thank to TUBITAK for their supports.
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical standards
Ethical approval for the study was granted by the ethics committee of Hacettepe University, Ankara, Turkey (GO 22/807). Informed consent from all participants was obtained.









