Introduction
Aplectana hylambatis (Baylis, 1927) is a cosmocercid (Cosmocercoidea: Cosmocercidae) parasite common to anurans from Argentina, Ecuador, Guyana, Paraguay, Peru, Uruguay, and Brazil (Campião et al., Reference Campião, Morais, Dias, Aguiar, Toledo, Tavares and Da Silva2014, Reference Campião, da Silva, Dalazen, Paiva and Tavares2016; Aguiar et al., Reference Aguiar, Honorio, Silva and Da Silva2015). This species has apparent low host specificity because it has been reported to infect anurans from different families (Campião et al., Reference Campião, Morais, Dias, Aguiar, Toledo, Tavares and Da Silva2014; Aguiar et al., Reference Aguiar, Honorio, Silva and Da Silva2015; González et al., Reference González, Gómez and Hamann2019). Although the morphology of A. hylambatis has been well-studied (González et al., Reference González, Gómez and Hamann2019), the species is not genetically characterised yet. In fact, from the 58 valid species of Aplectana (Santos et al., Reference Santos, Borges, Willkens, Santos, Costa-Campos and Melo2023), only four are available in GenBank database, in addition to others not identified to species level.
During parasitological studies on Pithecopus azureus (Cope, 1868) (Anura: Hylidae), collected in the Pantanal wetlands, Brazil, nematode specimens were found parasitizing the large intestine. After detailed morphological evaluation, they were identified as A. hylambatis and submitted to molecular analysis. Here, we provide the first genetic characterisation for the species, along with its phylogenetic position in the Cosmocercidae. Moreover, we provide a clear specific diagnosis for the material used in the genetic studies.
Materials and Methods
Seventy-five specimens of Pithecopus azureus (snout-vent length 34.1 ± 5.2 [21.0–47.0] mm) were collected through active search, in the Nhecolândia region (Pantanal wetlands), municipality of Corumbá, State of Mato Grosso do Sul, Brazil (18°59’25’’S, 56°39’40’’W), from December 2014 to November 2017. Anurans were euthanized by an injection of pentobarbital (10 mg/kg) in the body cavity, and immediately analysed for parasites, according to standard parasitological procedures (Amato et al., Reference Amato, Boeger and Amato1991). Host collection, transport, and euthanasia were approved by the Sistema de Autorização em Biodiversidade (acronym SISBIO; license number 5739-1) and the Ethics Committee for Animal Use of the Universidade Federal de Mato Grosso do Sul (acronym CEUA-UFMS, license number 878/2017).
Nematodes found alive infecting the large intestine were cleaned in saline, fixed in hot 4% formaldehyde and stored in 70% ethanol, for morphological evaluation. Four male specimens from different infrapopulations (see Bush et al., Reference Bush, Lafferty, Lotz and Shostak1997), had their midbody parts excised, fixed in 100% molecular biology grade ethanol, and stored at –22 °C for DNA isolation; the anterior and posterior parts of each specimen were processed for morphological studies, as previously described. Parasites, including the body parts of the males from which tissue was taken for genetic characterisation, were deposited in the Coleção Zoológica da Universidade Federal de Mato Grosso do Sul (acronym ZUFMS; accession numbers ZUFMS-AMP12210, 12221, 1231). The ecological terminology used herein follows Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997).
Morphological observations of the nematodes were performed using light and scanning electron microscopy (SEM) as described by Pereira et al. (Reference Pereira, Ferreira, Tomas, Elisei, Paiva and Tavares2019), and their specific identification was based on González et al. (Reference González, Gómez and Hamann2019).
DNA was isolated from small tissue samples, placed in a microcentrifuge sterile tube (1.5 mL). The sample was cut into small fragments with a sterile scissor, then we added 195 μL of 5% Chelex 100 (Sigma-Aldrich, Saint Louis, Missouri, USA) and 5 μL of proteinase K (20 mg/mL) (Phoneutria, Minas Gerais, Brazil). The tube was briefly vortexed, centrifuged at 3000 rpm, and incubated at 60 °C for digestion. Vortex and centrifugation steps were repeated from time to time, until the complete tissue digestion (~2 h). Digestion was followed by an enzymatic denaturation at 95 °C for 10 min. The tube was vortexed, centrifuged at 13,000 rpm for 10 min, and the supernatant containing DNA was stored at –22 °C.
Polymerase chain reactions (PCRs) and cycling conditions were the same described by Ailán-Choke et al. (Reference Ailán-Choke, Davies, Malta, Couto, Tavares, Luque and Pereira2023), using the primers Nema18SF + Nema18SR (Floyd et al., Reference Floyd, Rogers, Lambshead and Smith2005) that flank the 5′ end of the 18S rDNA, and D2A + D3B (De Ley et al., Reference De Ley, De Ley, Morris, Abebe, Mundo-Ocampo, Yoder, Heras, Waumann, Rocha-Olivares, Jay Burr, Baldwin and Thomas2005) that flank the domains D2A and D3B of the 28S rDNA. PCR products were visualized using agarose gel electrophoresis; the positive amplicons were submitted to enzymatic treatment with ExoSAP-IT (Thermo Fisher, Massachusetts, USA), and sent for Sanger sequencing at ACTGene (Ludwig Biotec, Rio Grande do Sul, Brazil).
Contig sequences were visualised, assembled, edited for primer trimming, and the consensus extracted in Geneious Prime (Dotmatics). We used the BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the close relationship between the sequences obtained for A. hylambatis and those of cosmocercid nematodes. Based on the BLAST results, we selected sequences from GenBank with the objective of evaluate the phylogenetic position of A. hylambatis in the family Cosmocercidae, based on the following criteria: sequences that covered the same genetic region as those from the present study, identified to species level and present in scientific publications. Unfortunately, there are few 28S sequences of cosmocercid nematodes that overlap those obtained here, in which the overlapping fragment is about 455-bp long. Therefore, we decided to work on two datasets: one consisting of 18S sequences and one consisting of 18S + 28S sequences. It proportionated the best taxa coverage and the evaluation of the phylogenetic informativeness associated with 18S and 28S. Sequences were aligned using MUSCLE, with default options, in Geneious Prime. Alignments were not edited, only trimmed in both ends to equalize sequence size. The reliability of the aligned positions was inspected using TCS (Chang et al., Reference Chang, Di Tommaso, Lefort, Gascuel and Notredame2015). Sequences used in the phylogenetic analyses are in the Supplementary Table S1.
Phylogenies were inferred using Bayesian inference in BEAST2 (Bouckaert et al., Reference Bouckaert, Vaughan, Barido-Sottani, Duchêne, Fourment, Gavryushkina, Heled, Jones, Kühnert, De Maio, Matschiner, Mendes, Müller, Ogilvie, du Plessis, Popinga, Rambaut, Rasmussen, Siveroni, Suchard, Wu, Xie, Zhang, Stadler and Drummond2019). The best-fit nucleotide substitution model and its parameters for each dataset (partitioned in the 18S + 28S), was evaluated using dModelTest (Bouckaert & Drummond, Reference Bouckaert and Drummond2017), implemented in BEAST2. The molecular clock model was relaxed (log exponential), defined according to the nested sampling method (Russel et al., Reference Russel, Brewer, Klaere and Bouckaert2019) and the Yule tree prior, which was selected according to the posterior densities and the effective sample sizes (ESS), calculated in Tracer (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018). The posterior estimates of parameter densities and the ESS for each parameter, as well as the posterior probability for branch supports in the majority rule consensus phylogenetic trees, were determined after running the Markov chain Monte Carlo, in two runs of four chains, each run with 20 × 106 generations, 25% of burn-in and saving the last 10,001 trees. The quality of the analysis (parameter densities, ESS, and burn-in) and the chain convergence were examined in Tracer (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018). Dichelyne grandistomis (Ferraz & Thatcher, 1988) was used as outgroup, based on the phylogenetic reconstruction of nuclear ribosomal sequences provided by Choudhury & Nadler (Reference Choudhury and Nadler2016).
The phylogeny and the alignment of 18S + 28S sequences were subjected to an analysis of phylogenetic informativeness using PhyDesign (López-Giráldez & Townsend, Reference López-Giráldez and Townsend2011), in the webserver http://phydesign.townsend.yale.edu/.
Results and discussion
Prevalence, mean intensity, and mean abundance of parasite infections were 5.3% (4/75), 12 ± 11.6 (2–26), and 0.64 ± 3.57 (0–26), respectively. All parasites were found in the large intestine and in adult stage.
We observed the following morphological features that were used for species diagnosis. The males had a total of 13 pairs of caudal papillae: three pairs were subventral to sublateral and precloacal; two pairs were laterally displaced at cloacal region; three pairs were slightly anterior to the cloacal opening; five pairs were postcloacal, in which the first two pairs were adjacent and subventral, followed by one pair sublateral at the mid-length of the tail, and two pairs at the same level, near the tail tip, of which one was ventral and one was lateral; plus one large unpaired papilla at the anterior cloacal labium. The spicules had a distal articulation that may give them a sickle shape, when protracted from the body; this feature was better observed using SEM. The females had a prominent vulva and, in the vulvar region, there were two conspicuous mamelon-like structures, both anterior to the vulva, in which one was laterally displaced. The features described here, along with a general view of the morphology observed in the present specimens, are depicted in Supplementary Figures S1–S9.
The present specimens showed morphometric features that were within the range of variation observed for A. hylambatys. However, some males and females were slightly smaller which also resulted in slightly smaller anatomic structures. We provide the comparative morphometry of A. hylambatis, including the present data and that from previous taxonomic studies of the species, in Supplementary Table S2.
There was no genetic divergence in the sequences obtained from the different male specimens, for both genetic markers. Therefore, only one 18S and one 28S sequence was used in the phylogenetic reconstructions. The lengths of the 18S and 28S sequences were 805 bp and 758 bp, respectively. The BLAST search indicated a genetic similarity of 99.0% between the present 18S sequence and the one labelled as Raillietnema sp. (DQ503461). Moreover, 18S genetic similarity between the present sample, and those of A. chamaeleonis (Baylis, 1929) and Cosmocerca ornata (Dujardin, 1845) was 98.3%. Regarding the 28S, the newly obtained sequence was more similar to those of C. ornata, Cosmocercoides tonkinensis Tran, Sato & Luc, 2015 and Co. pulcher Wilkie, 1930 (genetic similarity ranging from 92.1% to 92.5%).
The alignment of the 18S sequences had 808 sites, of which 101 were variable and corresponded to 12.5% of the aligned positions. The alignment of the 18S and the 28S sequences concatenated had 1,265 sites, of which 323 were variable and corresponded to 25.5% of the aligned positions. Sequence lengths of the samples were the same in all genetic alignments. The best-fit substitution model for the 18S alignments (partitioned in the combined dataset) was the TN93, with equal base frequencies and three different categories of nucleotide substitution; that for the 28S (partitioned in the combined dataset) alignment was also TN93, but with unequal base frequencies and four different categories of nucleotide substitution. All the original files including genetic alignments, outputs of dModelTest and BEAST2, as well as the raw phylogenetic trees are provided in Supplementary Material S1.
The phylogenetic reconstructions based on 18S and 18S + 28S sequences, generated similar topologies, in which nodal supports were improved when using both genes (Figure 1). Only the phylogenetic relationships with high nodal supports will be addressed here. According to both phylogenies, Aplectana was not monophyletic because two species of Cosmocerca clustered with it (Figure 1). Aplectana hylambatis formed a basal lineage within the group formed by its congeners, and Cosmocerca spp. (Figure 1). Aplectana chamaeleonis was sister to C. simile Chen, Zhang, Feng & Li, 2020, and the group formed by them was sister to C. ornarta (Figure 1). Aplectana xishuangbannaensis Chen, Gu, Ni & Li, 2021 appeared as a sister group of A. dayaoshanensis Chen, Ni, Gu, Sinsch & Li, 2021. The genus Cosmocercoides, was monophyletic and sister to Nemhelix bakeri Morand & Petter, 1986 in the phylogeny inferred from 18S sequences (Figure 1). The phylogenetic informativeness profile of 28S was higher than that of 18S sequences, in which there was a sudden drop on it, regarding the most recent divergence events (Figure 1).
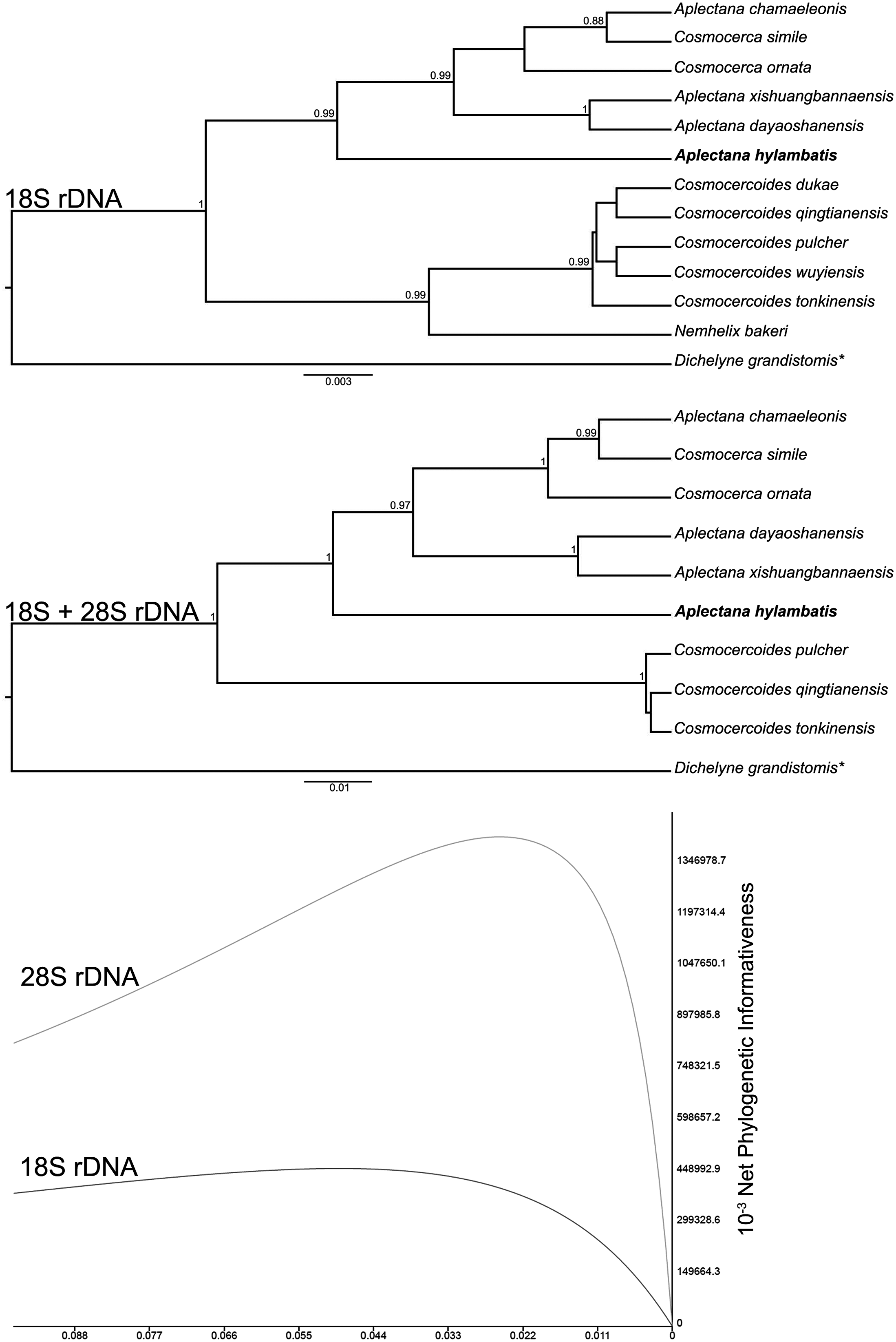
Figure 1. Phylogenetic reconstructions using Bayesian inference, from genetic sequences of cosmocercid nematodes. Nodal supports lower than 0.85 were omitted. Scale bars indicate substitution per site. The sample of the present study is in bold. The graph at the bottom indicates the phylogenetic informativeness profile of 18S and 28S rDNA genetic markers and is related to the tree immediately above.
The present prevalence and mean abundance of A. hylambatis in P. azureus can be considered low in comparison with conspecific infections in hosts from different families, other than Hylidae (Aguiar et al., Reference Aguiar, Honorio, Silva and Da Silva2015; Campião et al., Reference Campião, da Silva, Dalazen, Paiva and Tavares2016; González et al., Reference González, Gómez and Hamann2019). This parasite has been reported more frequently in anurans from Bufonidae, Leptodactylidae and Microhylidae (Campião et al., Reference Campião, Morais, Dias, Aguiar, Toledo, Tavares and Da Silva2014, Reference Campião, da Silva, Dalazen, Paiva and Tavares2016). Although P. azureus or other hylids may not be the most suitable hosts for A. hylambatis, this is the first report of A. hylambatis infecting P. azureus.
González et al. (Reference González, Gómez and Hamann2019) provided the most comprehensive morphological analysis of A. hylambatis. According to these authors, the species may show both morphological and morphometric variations, which are accounted by differences on host species and geographic distribution. Despite such an intraspecific morphological variability, the presence of an articulation at the distal portion of the spicules, which gives it a sickle shape when protruded for the body, and one to three conspicuous mamelon-like structures slightly anterior to the vulva, seem to be important for the diagnosis of A. hylambatis. These features were present in all of the observed parasites, and the mamelon-like structures occurred in pairs. It should be mentioned that the spicules may not assume a sickle shape under light microscopy, depending on the position of the specimen; however, it is easily visible using SEM. Moreover, it is possible to see the internal articulation zone at the distal end of the spicule, which appears in the form of a groove.
The number and arrangement of caudal papillae in males are commonly used for the diagnosis of nematode parasite species. In A. hylambatis they present some variation (González et al., Reference González, Gómez and Hamann2019), but still have the following features that seem to be frequent: three pairs and an unpaired papilla slightly anterior to the cloaca, of which one pair is anteriorly displaced; two pairs of lateral papillae slightly posterior and anterior to the cloacal opening, respectively, or somewhat at the same level of it.
All variations attributed to A. hylambatis, as well as its low host specificity and wide geographic distribution, may suggest that it is not a single species, but a species complex. Such a similar situation was observed for P. retusa, a widely distributed nematode, parasite in different species of reptiles and amphibians (Ailán-Choke et al., Reference Ailán-Choke, Ferreira, Paiva, Tavares, Paschoal and Pereira2024). In this sense, this first genetic characterisation of A. hylambatis would be useful for further studies on the population genetics of this taxon.
The sequences obtained in the present study were close to those of cosmocercid nematodes, according to the BLAST search and the phylogenetic reconstructions. Moreover, A. hylambatis was sister to the other congeners. These results confirm the generic assignment and its morphological identification. However, Aplectana was not monophyletic, because C. simile and C. ornate was grouped with Aplectana spp. It is interesting that almost all Aplectana and Cosmocerca samples originate from China, but A. chamaeleonis that were grouped with C. simile originates from Germany. In addition, the present sample from Brazil was basal in the clade formed by species of Aplectana and Cosmocerca. These results indicate some degree of geographic influence on the divergence processes of these taxa, which placed the Brazilian species more distant from those of Europe and China.
According to Chen et al. (Reference Chen, Gu, Ni and Li2021a), Aplectana and Cosmocerca are closer to each other than to Cosmocercoides. Furthermore, Chen et al. (Reference Chen, Ni, Gu, Sinsch and Li2021b) and Svitin et al. (Reference Svitin, Kuzmin, Harnoster, Net and du Preez2023) observed a lack of monophyly in Aplectana, due to the presence of Cosmocerca spp. that grouped together with it. The present findings corroborate these results. Therefore, it is possible that some of the diagnostic features of certain genera in Cosmocercinae are homoplastic; for example, the presence of plectanes and of caudal papillae on the plectanes which are used to differentiate Aplectana from Cosmocerca (Anderson et al., Reference Anderson, Chabaud and Willmott2009). However, such a discussion will require evidence based on a more diverse genetic database and integrative taxonomic approaches. In fact, it is hard to discuss the phylogeny of Cosmocercidae today, due to the low diversity of genetic sequences available. Several genera and species in this family still lack genetic characterisation, a fact that reinforces the importance of the present results.
Although the 28S alignment was shorter, it was more phylogenetically informative than that of 18S. Therefore, it is reasonable to suggest that the 28S may be considered important in assessing the relationships at higher and intermediate levels, among cosmocercid nematodes. 18S and 28S are not as suitable for diagnosing species as internal transcribed spacer or mitochondrial markers. However, our objective was to evaluate the phylogenetic position of A. hylambatis in the Cosmocercidae which clearly indicated that the species is distant from its Old World and Eastern congeners. Moreover, this first genetic characterisation for the species, especially regarding the 28S sequence, will be important for future evaluations of A. hylambatis from different hosts and geographic origins, since the 28S D2A-D3B region has been shown to be useful in population genetic studies of nematode parasites (Nadler & Pérez-Ponce de Leon, Reference Nadler and Pérez-Ponce de Leon2011; Pereira & Luque, Reference Pereira and Luque2017).
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/S0022149X24000609.
Acknowledgements
F.B.P. was supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Brazil (FAPEMIG; Process: APQ-01179-21). P.S.d.S. was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq; Process: 151858/2022-5).
Competing interest
None.
Ethical standard
All procedures involving animals were strictly in accordance with the ethical standards for animal use and approved by SISBIO (license number 57379-1) and CEUA (process number 878/2017) (Ethics Committee for Animal Use).



