Introduction
Adults of the genus Maritrema Nicoll, 1907 are intestinal endoparasites of aquatic birds of the families Laridae, Scolopacidae, Ardeidae, Threskiornithidae, and sporadically of passerine birds and mammals, distributed worldwide (Deblock Reference Deblock1971; Tkach Reference Tkach1988; Presswell et al. Reference Presswell, Blasco-Costa and Kostadinova2014; Hernández-Orts et al. Reference Hernández-Orts, Capasso, Pinacho-Pinacho and García-Varela2020). Morphologically, the genus is characterized by the presence of a small and linguiform body covered with tiny spines, a follicular vitellarium in two symmetrical ribbons surrounding uterus and testes, a small unspecialized genital atrium, and a characteristic postcecal position of the uterine coils (Deblock Reference Deblock1971, Reference Deblock, Bray, Gibson and Jones2008). Of the 70 described species of Maritrema, only a few (14 species representing 20% of the biodiversity) have been sequenced to delineate species and to understand their phylogenetic relationships among the family Microphallidae Travassos, 1920. Analyses inferred with the large subunit from nuclear DNA show the genus Maritrema as a monophyletic assemblage and sister group of the genus Microphallus Ward, 1901 (Galaktionov et al. Reference Galaktionov, Blasco-Costa and Olson2012; Presswell et al. Reference Presswell, Blasco-Costa and Kostadinova2014; Kudlai et al. Reference Kudlai, Cutmore and Cribb2015, Reference Kudlai, Cribb and Cutmore2016; Hernández-Orts et al. Reference Hernández-Orts, Pinacho-Pinacho, García-Varela and Kostadinova2016, Reference Hernández-Orts, Capasso, Pinacho-Pinacho and García-Varela2020; Blasco-Costa et al. Reference Blasco-Costa, Seppälä, Feijen, Zajac, Klappert and Jokela2020; Quinn et al. Reference Quinn, Thomas, Malkin, Eley, Coates and Rowley2022; Sokolov et al. Reference Sokolov, Shchenkov, Khasanov and Gordeev2023).
In the neotropical region of Mexico, adults of three species of Maritrema have been recorded in the coasts of the Pacific Ocean and Gulf of Mexico – that is, M. patulus Coil, Reference Coil1955 from the solitary sandpiper Tringa solitaria (Wilson) in Oaxaca state, M. corai Hernández-Orts, Pinacho-Pinacho, García-Varela & Kostadinova, Reference Hernández-Orts, Pinacho-Pinacho, García-Varela and Kostadinova2016 from the white ibis Eudocimus albus (L.) in Nayarit and Guerrero, and M. kostadinovae Hernández-Orts, Capasso, Pinacho-Pinacho & García-Varela, Reference Hernández-Orts, Capasso, Pinacho-Pinacho and García-Varela2020 from the yellow-crowned night heron Nyctanassa violacea (L.) in Veracruz (Coil Reference Coil1955; Hernández-Orts et al. Reference Hernández-Orts, Pinacho-Pinacho, García-Varela and Kostadinova2016, Reference Hernández-Orts, Capasso, Pinacho-Pinacho and García-Varela2020). In the last years, we prospected birds of three species distributed sympatrically in the Gulf of Mexico – namely, Tringa semipalmata Gmelin (Scolopacidae), Leucophaeus atricilla Linnaeus (Laridae), and Buteogallus urubitinga Gmelin (Accipitridae). Gravid specimens of microphallids belonging to the genus Maritrema were collected from the intestine of these species. The morphological examination of the specimens in combination with DNA sequence data and ecological information revealed they represent two undescribed species. The objectives of the present research were (1) to describe two new species of Maritrema and (2) to test their systematic position within Maritrema by using sequences of the large subunit (LSU) of the nuclear DNA.
Materials and methods
Seven birds of three species were hunted in three localities of southeastern Mexico: great black hawks (n=3) (B. urubitinga) in Tupilco, Tabasco (18° 26’ 6.004’’ N; 93° 7’ 44.60’’ W), laughing gull (n=2) (L. atricilla) in Isla Aguada, Campeche (18° 48’ 22.92’’ N, 91° 28’ 03.68’’ W), and the willet (n=2) (T. semipalmata) in Progreso, Yucatán (21° 13’ 37.888’’ N, 89° 49’ 40.252’’ W) (Figure 1). Birds were identified following Howell and Webb (Reference Howell and Webb1995) and the American Ornithologists’ Union (1998) guidelines. Adult digeneans were removed from the intestine of the birds, placed in Petri dishes with 0.6% saline solution, and examined under a stereomicroscope (EZ4 Leica). Microphallid specimens were relaxed in hot (near boiling) distilled water and preserved in 100% ethanol for morphological and molecular analyses.

Figure 1. Map of Mexico showing the sampled sites. Locality, 1 Tupilco, Tabasco (18° 26’ 6.004’’ N; 93° 7’ 44.60’’ W); Locality 2, in Isla Aguada, Campeche (18° 48’ 22.92’’ N, 91° 28’ 03.68’’ W); Locality 3, Progreso, Yucatán (21° 13’ 37.888’’ N, 89° 49’ 40.252’’ W); Locality 4, Salina Cruz, Oaxaca (16° 10’ 31.26’’ N, 95° 11’ 39.263’’ W); Locality 5, Laguna Chautengo, Guerrero (16° 38’ 07’’ N, 99° 08’ 26’’ W); Locality 6, La Tovara, Nayarit (21° 30’ 0’’ N, 105° 16’ 00’’ W); Locality 7, La Cortadura, Veracruz (22° 10’ 55.413’’ N, 98° 1’ 18.601’’ W); Locality 8, La Ribera, Veracruz (22° 6’ 54’’ N, 97° 46’ 37.999’’ W); Records, Maritrema corai, (![]() ); Maritrema itzamnai (
); Maritrema itzamnai (![]() ); Maritrema kukulkanni 2 (
); Maritrema kukulkanni 2 (![]() ); Maritrema patulus (
); Maritrema patulus (![]() ); Maritrema kostadinovae (
); Maritrema kostadinovae ( ); Red color, current records; Green color, previous records.
); Red color, current records; Green color, previous records.
Morphological analyses
Digeneans preserved in ethanol 100% were stained with Mayer’s paracarmine (Merck, Darmstadt, Germany), dehydrated in ethanol series, cleared in methyl salicylate, and mounted in Canada balsam for morphological analysis. Specimens were examined using a compound microscope equipped with a bright field Leica DM 1000 LED microscope (Leica, Wetzlar, Germany). Measurements were taken using Leica Application Suite microscope software (Leica Microsystems GmbH, Wetzlar, Germany) and are given in micrometers and presented with the range followed by the mean in parentheses. Some specimens were critical point dried with CO2, sputter coated with gold, and examined with a Hitachi Stereoscan Model S-2469N scanning electron microscope operating at 15 kV. Voucher specimens were deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México (UNAM), Mexico City.
Amplification and sequencing of DNA
Prior to extraction of the genomic DNA, each specimen was mounted on a microscope slide, and images were taken as references with a bright field Leica DM 1000 LED microscope (Leica, Wetzlar, Germany). Each image was linked with its genomic DNA, known as a photogenophore (see Andrade-Gómez and García-Varela Reference Andrade-Gómez and García-Varela2021) (Figure 2 a–c). Specimens were removed from the microscope slide, placed individually in a tube, and digested overnight at 56°C. The digestion solution contained 10 mM Tris-HCl (pH 7.6), 20 mM NaCl, 100 mM Na2 EDTA (pH 8.0), 1% sarcosyl, and 0.1 mg/ml proteinase K. Following digestion, DNA was extracted from the supernatant using DNAzol reagent (Molecular Research Center, Cincinnati, Ohio) according to the manufacturer’s instructions. The D1–D3 domains of the LSU of nuclear DNA were amplified using the forward 391, 5′-AGCGGAGGAAAAGAAACTAA-3′ (Nadler et al. Reference Nadler, Hoberg, Hudspeth and Rickard2000) and the reverse 536, 5′-CAGCTATCCTGAGGGAAAC-3′

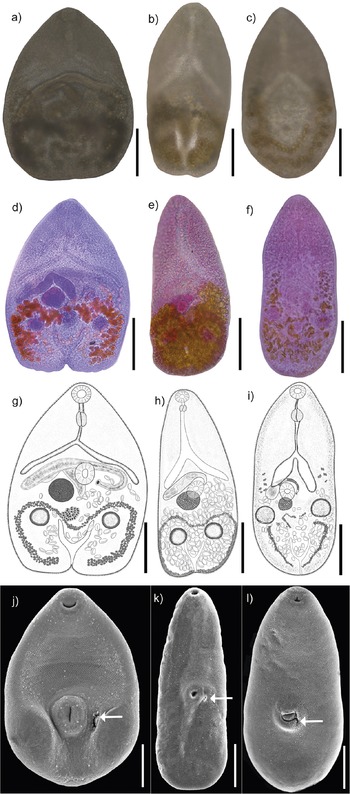
Figure 2. Photogenophores (a–c); Photographs of stained whole mounted specimnes (d–f); Drawings of complete adult specimens (g–i); Scanning electron photomicrographs (j–l) of Maritrema corai (a, d, g, j); Maritrema itzamnai n. sp. (b, e, h, k); Maritrema kukulkanni n. sp. (c, f, i, l); Arrows indicate the genital pore (j, k, l). Scale bars = 100 μm.
(García-Varela and Nadler Reference García-Varela and Nadler2005). PCRs (25 μl) consisted of 1 μl of each primer (10 μM), 2.5 μl of 10X PCR Rxn buffer, 1.5 μl of 2 mM MgCl2, 0.5 μl of dNTPs (10 mM), 16.375 μl of water, 2 μl of genomic DNA, and 1 U of Taq DNA polymerase (Platinum Taq, Invitrogen Corporation, São Paulo, Brazil). The PCR cycling parameters for rDNA amplification included denaturation at 94°C for 1 min, followed by 35 cycles at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min, with postamplification incubation at 72°C for 10 min. Sequencing reactions were performed using the initial primers plus two internal primers, 502 (5’–CAAGTACCGTGAGGGAAAGTTGC–3’) and 503 (5’– CCTTGGTCCGTGTTTCAAGACG–3’) (García-Varela and Nadler Reference García-Varela and Nadler2005), with ABI Big Dye (Applied Biosystems, Boston, Massachusetts) terminator sequencing chemistry, and reaction products were separated and detected using an ABI 3730 capillary DNA sequencer. Contigs were assembled and base-calling differences were resolved using Codoncode Aligner version 9.0.1 (Codoncode Corporation, Dedham, Massachusetts).
Alignments and phylogenetic analyses
LSU sequences obtained in the current research were aligned separately with 16 other sequences representing 14 species of Maritrema downloaded from the GenBank database, plus seven sequences from the genus Microphallus and Plagiorchis vespertilionis (Müller, 1780) Barun, 1900 (GenBank No. AF1511931), Telorchis assula (Dujardin, 1845) (AF151915), and Haematoloechus longiplexus (Stafford, 1902) (AF387801), used as outgroups. The alignment was performed using the software Clustal W (Thompson et al. Reference Thompson, Higgins and Gibson1994) with default parameters and adjusted manually with the software Mesquite (Maddison and Maddison Reference Maddison and Maddison2011). The alignment consisted of 33 sequences with 1,318 nucleotides. The best nucleotide substitution model was estimated with the Akaike information criterion (AIC) implemented in jModelTest v0.1.1 (Posada Reference Posada2008). The phylogenetic analyses were performed using maximum likelihood (ML) and Bayesian inference (BI) methods. ML was carried out with RAxML version 7.0.4 (Silvestro and Michalak Reference Silvestro and Michalak2011), and Bayesian inference (BI) analyses were performed with MrBayes version 3.2.7 (Ronquist et al. Reference Ronquist, Teslenko, Van Der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) using the Cyberinfrastructure with Phylogenetic Research (CIPRES) Science Gateway v3.3 online interface (Miller et al. Reference Miller, Pfeiffer and Schwartz2010). To support each node, 10,000 bootstrap replicates were run with ML. BI analyses included Markov chain Monte Carlo (MCMC) searches with two simultaneous runs for 10 million generations, sampling every 1,000 generations, a heating parameter value of 0.2, and a “burn-in” of 25%. Trees were visualized in FigTree v.1.3.1 (Rambaut 2012). The genetic divergence among taxa was estimated using uncorrected “p” distances with the program MEGA version 11 (Tamura et al. Reference Tamura, Stecher and Kumar2021).
Results
Taxonomy
Family: Microphallidae Ward, 1901.
Subfamily: Maritrematinae Nicoll, 1909.
Genus: Maritrema Nicoll, 1907.
Maritrema corai Hernández-Orts, Pinacho-Pinacho, García-Varela & Kostadinova, Reference Hernández-Orts, Pinacho-Pinacho, García-Varela and Kostadinova2016.
Type host: white Ibis, Eudocimus albus (Linnaeus, 1758).
Other host: Buteogallus urubitinga (Gmelin, 1788).
Site of infection: Small intestine.
Prevalence: 33.3% (1/3).
Type locality: La Tovara, San Blast, Nayarit (21° 32’ 0’’ N, 105° 16’ 00’’ W).
Other localities: Chautengo, Guerrero (16° 38’ 07’’ N, 99° 08’ 26’’ W); Tupilco, Tabasco (18° 26’ 6.004’’ N; 93° 7’ 44.60’’ W, this work).
Voucher: CNHE: 11914.
Representative DNA sequences: PP541428, PP541429 (LSU).
Morphological identification
Five adult specimens were collected, measured, and compared with described species. Our specimens showed similar morphological characteristics compared with those assigned to M. corai by Hernández-Orts et al. (Reference Hernández-Orts, Pinacho-Pinacho, García-Varela and Kostadinova2016), including a pyriform body covered with minute spines; an oral sucker subterminal, subspherical, an intestinal bifurcation in anterior third of body; a J-shaped cirrus sac; a sinistrolateral seminal vesicle; a dextral ovary; and vitellarium restricted to hindbody forming small follicles (Figure 2a, d, g, j; Table 1).
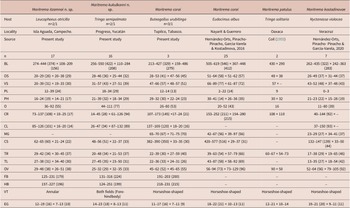
Table 1. Comparative measurements of Maritrema itzamnai n. sp., Maritrema kukulkanni n. sp. and its congeneric species recorded in coasts of Mexico. Abbreviations: BL, body; OS, oral sucker; VS, ventral sucker; PL, pre-pharynx; PH, pharynx; O, oesophagus; CR, caeca right; CL, caeca left; MG, Mehlis gland; CS, cirrus sac; TR, testes right; TL, testes left; OV, ovary; FB, forebody; HB, hindbody, VT, vitellarium shape; EG, egg

Maritrema itzamnai n. sp.
Type host: laughing gull, Leucophaeus atricilla (Linnaeus, 1758).
Site of infection: Small intestine.
Prevalence: 50% (1/2).
Type locality: Isla Aguada, Campeche (18° 48’ 22.92’’ N, 91° 28’ 03.68’’ W).
Type-material: CNHE: 11910 (holotype); 11911 (paratype).
Representative DNA sequences: PP541430, PP541431 (LSU).
Etymology: The specific epithet refers to Itzamná, the god of the sky in the Mayan civilization; Mayans inhabited southeastern Mexico for approximately 4,000 years.
Description
Maritrema itzamnai n. sp. (Figure 2b, e, h, k). (Based on 17 mounted specimens and 3 specimens from SEM). Body dorsoventrally flattened, pyriform with maximum width at level of ventral sucker 108–209 (156) (Figure 2e, h). Body length 274–444 (374). Tegument entirely covered with minute spines (Figure 2k). Forebody 125–231 (179) long. Oral sucker subterminal, subspherical, 20–29 × 20–38 (26 × 29). Ventral sucker equatorial, subspherical, 20–39 × 19–33 (31 × 30), larger than oral sucker; sucker length ratio 1: 0.87–1.0 (1:0.92); sucker width ratio 1:1.05–1.15 (1: 1.09). Prepharynx 12–39 (24) long. Pharynx small, subspherical, 16–24 × 14–21 (19 × 17). Oesophagus 36–92 (55) long, with epithelial lining. Intestinal bifurcation pre-equatorial, immediately anterior to cirrus sac. Caeca, short, with epithelial lining, divergent, reaching anterior or middle level of ventral sucker, right caecum 73–137 × 10–25 (108 × 17), left caecum 85–126 × 16–20 (101 × 14). Testes two, slightly oblique, postovarian, subspherical, right testis 29–42 × 30–45 (34 × 37), left testis 27–38 × 34–40 (31 × 30). Cirrus sac transverse, anterior of ventral sucker dorsally 62–65 × 21–24 (60 × 22); numerous prostatic cell surrounding curled the ejaculatory duct and unarmed cirrus. Seminal vesicle elongate. Ejaculatory duct long. Genital pore oval (Figure 2k), left to ventral sucker (not observed in Figure 2h). Ovary subspherical, adjacent to ventral sucker dorsally, 29–48 × 26–51 (38 × 38). Oviduct not observed. Mehlis’s gland not distinct. Laurer’s canal not observed. Eggs numerous, small, 12–19 × 7–13 (16 × 10). Vitellarium in hindbody, comprises numerous small follicles forming two symmetrical ribbons, confluent posteriorly, enclosing the testes. Excretory vesicle short, terminal, Y-shaped; stem short, arms reach to posterior level of testes; pore terminal (see Figure 2e, h, k; Table 1).
Maritrema kukulkanni n. sp.
Type host: willet, Tringa semipalmata (Gmelin, 1789).
Site of infection: Small intestine.
Prevalence: 50% (1/2).
Type locality: Progreso, Yucatán (21° 13’ 37.888’’ N, 89° 49’ 40.252’’ W).
Type-material: CNHE: 11912 (holotype); 11913 (paratype).
Representative DNA sequences: PP541432-434 (LSU).
Etymology: The specific epithet refers to Kukulkán, feathered serpent god in the Mayan civilization.
Description
Maritrema kukulkanni n. sp. (Figure 2c, f, i, l). (Based on 16 mounted specimens and 2 specimens from SEM). Body dorsoventrally flattened, pyriform with maximum width at level of ventral sucker 110–284 (208) (Figure 2f, i). Body length 256–550 (422). Tegument entirely covered with minute spines (Figure 2l). Forebody 131–316 (224) long. Oral sucker subterminal, subspherical, 28–46 × 25–44 (36 × 32). Ventral sucker post-equatorial, subspherical, 31–57 × 27–51 (43 × 39), larger than oral sucker; sucker length ratio 1: 0.8–0.91 (0.83); sucker width ratio 1: 0.86–0.92 (0.87). Prepharynx, 18–34 (29) long. Pharynx subspherical, 21–39 × 18–34 (32 × 29). Oesophagus 44–111 (77) long, with epithelial lining. Intestinal bifurcation pre-equatorial, immediately anterior to cirrus sac. Caeca, short, with epithelial lining, divergent, reaching anterior or middle level of ventral sucker, right caecum 14–45 × 61–126 (28 × 94), left caecum 26–47 × 87–132 (34 × 89). Testes two slightly oblique, postovarian, subspherical, right testis 20–48 × 21–53 (34 × 37), left testis 27–45 × 23–50 (35 × 31). Cirrus sac transverse, at level of ventral sucker. Numerous prostatic cell surrounding curled the ejaculatory duct and unarmed cirrus. Seminal vesicle elongate-oval. Ejaculatory duct short. Genital pore oval, left to ventral sucker (Figure 2l). Ovary subspherical, adjacent to ventral sucker dorsally, 25–32 × 32–35 (29 × 33). Oviduct not observed. Mehlis’s gland not observed. Laurer’s canal not observed. Eggs numerous, small, 14–23 × 8–13 (18 × 11). Vitellarium in hindbody, comprises numerous small follicles at level of the caecum and reaching posterior end to excretory vesicle level. Excretory vesicle short, terminal, Y-shaped; stem short, arms reach to posterior level of testes; pore terminal (see Figure 2f, i, l; Table 1).
Taxonomic remarks
With the inclusion of two new species to Maritrema, now it contains 72 species distributed worldwide, associated to aquatic birds and mammals (see Hernández -Orts et al. 2016, 2020; Capasso et al. Reference Capasso, D’Amico and Diaz2019). Kinsella and Deblock (Reference Kinsella and Deblock1997) proposed two species groups within the genus Maritrema: 1) group Eroliae, which contains species with cirrus covered partially or entirely with spines and 2) group Patulus, which contains species with an unarmed cirrus and a horseshoe-shaped vitellarium. The two new species possess an unarmed cirrus and are morphologically similar. However, Maritrema itzamnai n. sp. can be easily differentiated from Maritrema kukulkanni n. sp. by having annular vitellarium instead of a vitellarium dispersed in both hindbody and forebody. Additionally, the two species can be differentiated in terms of the size of some morphological traits, with Maritrema itzamnai n. sp. being smaller – for example, pharynx 16–24 × 14–21, in M. itzamnai n. sp. vs. 21–39 × 18–34 in M. kukulkanni n. sp.; ovary 29–48 × 26–51 in M. itzamnai n. sp. vs. 25–32 × 32–35 in M. kukulkanni n. sp.; and cirrus sac size 62–65 × 21–24 in M. itzamnai n. sp. vs. 48–56 × 22–37 in M. kukulkanni n. sp. (see Table 1).
Furthermore, Maritrema itzamnai n. sp. can be morphologically differentiated from M. corai, M. patulus, and M. kostadinovae species distributed in the coasts of Mexico by having a smaller oral sucker 20–29 × 20–38 vs. 28–64 × 47–62 in M. corai; 48 × 38 in M. patulus, and 26–49 × 31–44 in M. kostadinovae, and a smaller ventral sucker, 20–39 × 19–33 vs. 47–89 × 48–87 in M. corai; 57 in M. patulus, and 43–52 × 36–48 in M. kostadinovae (Table 1). Moreover, Maritrema itzamnai n. sp. also differs from the other species in having an annular vitellarium instead of a horseshoe-shaped vitellarium as in M. corai, M. patulus, and M. kostadinovae. The second new species, Maritrema kukulkanni n. sp., differs from their congeners distributed in coasts of Mexico in the oesophagus length which is 44–117 vs. 20–80 in M. corai, 16 in M. patulus, and 11–80 in M. kostadinovae. Finally, Maritrema kukulkanni n. sp. can be differentiated in the vitellarium shape, which is dispersed in both the hindbody and forebody, whereas in the other three species, vitellarium is arranged as a horseshoe shape.
Phylogenetic analyses and genetic divergence
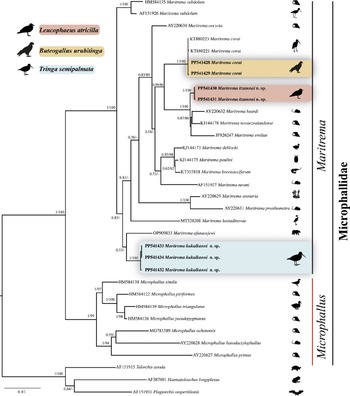
The phylogenetic trees recovered Maritrema as monophyletic with the highest bootstrap (100%) and Bayesian posterior probability support values (1.0) (Figure 3). The three species of Maritrema analysed in the current study were nested in three reciprocally monophyletic clades. The first clade was formed by two sequences of Maritrema itzamnai n. sp., which is sister to a clade formed by M. eroliae Yamaguti, 1939 (JF826247), which is sister to M. heardi (Kinsella & Deblock, Reference Kinsella and Deblock1997) Tkach, Littlewood, Olson, Kinsella & Swiderski, 2003 (AY220632) plus M. novaezealandense Presswell, Blasco-Costa & Kostadinova (KJ144178). The second clade was formed by three sequences of Maritrema kukulkanni n. sp., which is sister to M. afanassjewi Belopol’skaya 1952 (OP90983). Finally, the third clade was formed by two sequences of M. corai generated in this study plus other two isolates (KT880221 and KT880223) obtained from GenBank. The three clades were highly supported by bootstrap (100%) and Bayesian posterior probability values (1.0).

Figure 3. Maximum likelihood tree and consensus Bayesian Inference trees inferred with large subunit (LSU) from nuclear DNA; numbers near internal nodes show posterior probabilities (BI) and ML bootstrap values. Sequences in bold were generated in this study. Scale bar = number of substitutions; dash in the nodes represent values less than 50% of bootstrap.
The LSU intraspecific genetic divergence among four isolates of M. corai was very low, ranging from 0 to 0.2%, among three isolates of Maritrema kukulkanni n. sp. ranged from 0 to 0.2%, and among two isolates of Maritrema itzamnai n. sp., the sequences were identical. The interspecific divergence among Maritrema spp. varied between 1.5 and 8.4%; the greatest divergence was found between M. eroliae (JF826247) and M. afanassjewi (OP909833), and the lowest was found between M. novaezealandense (KJ144178) and M. heardi (AY220632).
Discussion
Members of the genus Maritrema are considered to be typical components of helminth fauna of brackish, marine, and, to a lesser extent, freshwater birds (Capasso et al. Reference Capasso, D’Amico and Diaz2019). The phylogenetic trees inferred with the LSU dataset unequivocally showed that Maritrema is monophyletic and shared a common ancestor with members of the genus Microphallus. This result is in agreement with previous phylogenetic studies that contributed significantly to our understanding of the systematics and classification of the family Microphallidae (Presswell et al. Reference Presswell, Blasco-Costa and Kostadinova2014; Kudlai et al. Reference Kudlai, Cribb and Cutmore2016; Hernández-Orts et al. Reference Hernández-Orts, Capasso, Pinacho-Pinacho and García-Varela2020; Blasco-Costa et al. Reference Blasco-Costa, Seppälä, Feijen, Zajac, Klappert and Jokela2020; Quinn et al. Reference Quinn, Thomas, Malkin, Eley, Coates and Rowley2022; Sokolov et al. Reference Sokolov, Shchenkov, Khasanov and Gordeev2023). Our phylogenetic analyses added new sequence data to a growing genetic library and allowed us to assess the phylogenetic relationships among the species of Maritrema. The ML and Bayesian analyses uncovered three independent subclades corresponding to three species, two of them new and described here. A detailed morphological study of all these specimens revealed unique morphological traits and synapomorphies shared with other members of Maritrema. In addition to morphological and molecular evidence, the genetic divergence found among the species studied provided additional information. For instance, interspecific genetic divergence among species of Maritrema ranged from 1.0 to 8.3%. Particularly, Maritrema itzamnai n. sp. diverged from its sister group composed by three species in 1.5% with respect to M. heardi, 1.0% with M. novaezealandense, and 2.3% with M. eroliae. The genetic divergence between Maritrema kukulkanni n. sp. and its sister species M. afanassjewi was 3.0%. The values of genetic divergence found are similar than estimated previously from 1.0 to 9.3% and from 5.25 to 7.92% among species of Maritrema (Presswell et al. Reference Presswell, Blasco-Costa and Kostadinova2014; Hernández-Orts et al. Reference Hernández-Orts, Capasso, Pinacho-Pinacho and García-Varela2020).
Of the 16 analyses species belonging to genus Maritrema, seven of them have been characterized genetically using larval stages, opening a window to link larval with adult stages, and even some unidentified isolates have been sequenced as part of a barcoding project (Pina et al. Reference Pina, Russell-Pinto and Rodrigues2011; Presswell et al. Reference Presswell, Blasco-Costa and Kostadinova2014). Currently, at least 32 species of Maritrema have been recorded in birds and mammals in the Americas. The complete life cycle of very few species of Maritrema is partially known in the Neotropical region (Díaz and Cremonte, 2010; Alda et al. Reference Alda, Bonel, Hechinger and Martorelli2013; Martínez et al. 2023). The current evidence indicates that adult worms of Maritrema spp. live and reproduce sexually in the digestive tract of aquatic birds that serve as definitive hosts. Eggs are expelled into the environment with the faeces of the definitive hosts. After the ingestion of the eggs by a cochliopid snail, Heleobia australis d’Orbigny, or by a pulmonate gastropod, Siphonaria lessonii Blainville, which serve as the first intermediate hosts, parasites develop into cercariae (Alda et al. Reference Alda, Bonel, Hechinger and Martorelli2013; Bagnato et al. Reference Bagnato, Gilardoni, Di Giorgio and Cremonte2015). The cercariae emerge and swim to find mainly crustaceans of the genera Neohelice Sakai, Türkay and Yang, and Cyrtograpsus Dana that act as second intermediate host, where they undergo encystment to develop into metacercariae. Finally, crustaceans with metacercariae are ingested by aquatic birds which are the definitive hosts (Díaz and Cremonte 2010; Alda et al. Reference Alda, Bonel, Hechinger and Martorelli2013; Capasso et al. Reference Capasso, D’Amico and Diaz2019; Martínez et al. 2023). The laughing gull (L. atricilla), the definitive host of Maritrema itzamnai n. sp., is an opportunistic forager, consuming both aquatic and marine invertebrates such as snails, earthworms, crabs, insects, mollusks, shrimps, and horseshoe crabs’ eggs (Davis Reference Davis2009). Moreover, the willet (T. semipalmata), which is the definitive host of Maritrema kukulkanni n. sp., feeds upon marine invertebrates, such as insects, crustaceans, mollusks, annelids, and occasionally fishes (Howell and Webb Reference Howell and Webb1995). Finally, the great black hawk (B. urubitinga), which is the definitive host of M. corai, feeds on small vertebrates (fish, amphibians, reptiles, birds, and mammals) and invertebrates (crabs and insects). Interestingly, the three birds analysed in this study feed off crustaceans (likely infected with metacercariae of Maritrema). In addition, the current record of M. corai from the great black hawk in Tupilco, Tabasco in the Gulf of Mexico (Locality 1, in Figure 1) represents a new locality record for M. corai, expanding its distribution range, since M. corai was originally described from the white ibis (Eudocimus albus) in the Pacific coasts of Mexico (localities 5 and 6 in Figure 1) (see Hernández-Orts et al. Reference Hernández-Orts, Pinacho-Pinacho, García-Varela and Kostadinova2016).
Authors’ contribution
YAP, MGV, GPPL, and BMG: conceptualization, phylogenetic analysis, writing – original draft, review, editing. YAP, MTGG, and MGV: sampling, investigation, genetic data, morphological study. MGV and GPPL: funding acquisition, review, and editing.
Acknowledgements
We sincerely thank Laura Márquez and Nelly López (LaNabio, Biology Institute, UNAM) for their help with DNA sequencing processing. A special thanks to Dra. Alejandra López Jimenez and Dr. Leopoldo Andrade Gómez for their help during the fieldwork.
Financial support
This research was supported by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM) IN201122 and IN200824 to MGV and GPPL, respectively.
Competing interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
Ethical standard
The sampling in this work complies with the current laws and animal ethics regulations of México.