Introduction
Peltigerales is one out of nine named orders in the most species-rich class among the ascomycetes, the Lecanoromycetes (Schoch et al. Reference Schoch, Sung, López-Giráldez, Townsend, Miądlikowska, Hofstetter, Robbertse, Matheny, Kauff and Wang2009), and incorporates the majority of lichen-forming fungi with cyanobacteria as their photosynthesizing symbiotic partner. The Peltigeralean lichens play an important role in the terrestrial nitrogen cycle of many ecosystems through the fixation of atmospheric nitrogen (Cleveland et al. Reference Cleveland, Townsend, Schimel, Fisher, Howarth, Hedin, Perakis, Latty, Von Fischer and Elseroad1999; Belnap Reference Belnap2003). Current classifications of the Peltigerales include ten families (Wedin et al. Reference Wedin, Jørgensen and Wiklund2007; Spribille & Muggia Reference Spribille and Muggia2013), four of which include c. 90% of the total number of species in the order, that is, Lobariaceae, Pannariaceae, Collemataceae, and Peltigeraceae (Kirk et al. Reference Kirk, Cannon, Minter and Stalpers2008). Several recent contributions have significantly increased knowledge about broad phylogenetic relationships in the Peltigerales (Wedin et al. Reference Wedin, Jørgensen and Wiklund2007, Reference Wedin, Wiklund, Jørgensen and Ekman2009; Otálora et al. Reference Otálora, Aragón, Molina, Martínez and Lutzoni2010; Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011; Spribille & Muggia Reference Spribille and Muggia2013).
Current estimates indicate that the Pannariaceae is the second most species-rich family of the Peltigerales, and includes more than 300 known species (Kirk et al. Reference Kirk, Cannon, Minter and Stalpers2008). In its original description (Tuckerman Reference Tuckerman1872), however, the Pannariaceae included only two genera, Pannaria and Heppia. It was not until the treatment by Zahlbruckner (Reference Zahlbruckner, Engler and Prantl1926) that the familial circumscription was stabilized and came to include large and well-known genera such as Psoroma and Parmeliella, which are still treated in the Pannariaceae. Zahlbruckner included eleven genera altogether, although he excluded Heppia. Some genera included by Zahlbruckner, Hydrothyrea, Massalongia, Placynthium and Coccocarpia, have later been excluded from the Pannariaceae (see, e.g. Wedin et al. Reference Wedin, Jørgensen and Wiklund2007, Reference Wedin, Wiklund, Jørgensen and Ekman2009). Jørgensen (Reference Jørgensen1978, Reference Jørgensen1994) pointed out that Zahlbruckner's generic classification had paid too much attention to the photobiont (green-algal or cyanobacterial) and presence or absence of a thalline margin in the apothecia. In the survey by Henssen & Jahns (Reference Henssen and Jahns1973), only four genera were included in the Pannariaceae: Lepidocollema, Pannaria, Parmeliella, and Psoroma. A preliminary single-gene phylogeny of the family (Ekman & Jørgensen Reference Ekman and Jørgensen2002) confirmed that Protopannaria is distinct from Pannaria (in which it had previously been included), that Pannaria included a mixture of species with a green-algal and cyanobacterial photobiont, and excluded the Fuscopannaria leucophaea group, later described as Vahliella (Jørgensen Reference Jørgensen2008), from the Pannariaceae. Continued revision of familial and generic boundaries led Jørgensen (Reference Jørgensen2003) to recognize 17 genera altogether, although some with doubt. Later investigations demonstrated that all studied genera with non-septate ascospores (Leciophysma, Physma, Ramalodium, and Staurolemma), traditionally referred to the Collemataceae because of their gelatinous thallus, should be transferred to the Pannariaceae (Wedin et al. Reference Wedin, Wiklund, Jørgensen and Ekman2009; Otálora et al. Reference Otálora, Aragón, Molina, Martínez and Lutzoni2010; Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011). In addition, Vahliella was shown to belong in a family of its own, Vahliellaceae (Wedin et al. Reference Wedin, Wiklund, Jørgensen and Ekman2009, Reference Wedin, Jørgensen and Ekman2011), whereas species with a Scytonema photobiont previously treated in Polychidium belong in a genus of Pannariaceae, Leptogidium (Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011).
Despite previous efforts, phylogenetic relationships within the Pannariaceae remain insufficiently known. Our aim was to estimate phylogenetic relationships in the Pannariaceae based on an expanded sampling of taxa, and provide a revised taxonomic overview of the family in light of the phylogenetic estimate, previous phylogenetic estimates, as well as morphological data.
Material and Methods
Taxonomy and nomenclature
We studied the type species of most described genera in the Pannariaceae, located in the herbaria cited in the taxonomic section below. The morphology and anatomy of the specimens were examined, and chemistry was investigated by thin-layer chromatography (Culberson & Kristinsson Reference Culberson and Kristinsson1970).
Taxon selection for molecular studies
We selected representatives of all genera included in the Pannariaceae as circumscribed by Jørgensen (Reference Jørgensen2003), Wedin et al. (Reference Wedin, Wiklund, Jørgensen and Ekman2009), Muggia et al. (Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011), and Spribille & Muggia (Reference Spribille and Muggia2013) except Kroswia (Jørgensen & Gjerde Reference Jørgensen and Gjerde2012), Leptogidium (Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011), Psoromidium (Galloway & James Reference Galloway and James1985), and Steineropsis (Spribille et al. Reference Spribille, Pérez-Ortega, Tønsberg and Schirokauer2010; Spribille & Muggia Reference Spribille and Muggia2013). We were unable to obtain fresh enough material of Lepidocollema and Psoromidium, whereas repeated attempts to generate PCR products from Kroswia were unsuccessful. Leptogidium and Steineropsis were not included because they were recognized as members of the Pannariaceae only after the initiation of this study (Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011; Spribille & Muggia Reference Spribille and Muggia2013). Altogether, the data matrix included 110 ingroup terminals representing 88 species (see Supplementary Materials , available online). Vahliella leucophaea, a member of the Vahliellaceae (Wedin et al. Reference Wedin, Jørgensen and Ekman2011), was used as outgroup.
DNA extraction, PCR amplification and sequence editing
We obtained DNA sequences from three different genes, the largest subunit of the RNA polymerase II gene (RPB1), the internal transcribed spacer (ITS) region (including ITS1, 5.8S, and ITS2) of the nuclear ribosomal RNA gene, and the small subunit of the mitochondrial ribosomal RNA gene (mrSSU). Laboratory methods followed Lindblom & Ekman (Reference Lindblom and Ekman2005), Ekman et al. (Reference Ekman, Andersen and Wedin2008), Wedin et al. (Reference Wedin, Wiklund, Jørgensen and Ekman2009), and Ekman & Blaalid (Reference Ekman and Blaalid2011).
Alignment of ITS
The ITS1 region was assumed to start immediately after the GATCATTA pattern at the end of the small subunit of the nuclear ribosomal RNA gene region. The ITS2 region was assumed to end after the 9th nucleotide preceding the TCGGATCA pattern at the beginning of the large subunit of the nuclear ribosomal RNA gene region. Borders between ITS1 and 5.8S and between 5.8S and ITS2 were defined using the Rfam 5.8S seed alignment (Gardner et al. Reference Gardner, Daub, Tate, Nawrocki, Kolbe, Lindgreen, Wilkinson, Finn, Griffiths-Jones and Eddy2009). A preliminary alignment was created using the G-INS-I algorithm of MAFFT version 6.820 (Katoh & Toh Reference Katoh and Toh2008). The ITS region was subsequently split into separate data sets. The 5.8S region was considered unambiguously and finally aligned, whereas the ITS1 and ITS2 regions were prepared for downstream structural alignment by stripping all gaps introduced by the preliminary alignment procedure. The two gene regions were subsequently aligned separately using three different structural aligners, viz. Murlet version 0.1 (Kiryu et al. Reference Kiryu, Tabei, Kin and Asai2007), CentroidAlign version 1.0 (Hamada et al. Reference Hamada, Sato, Kiryu, Mituyama and Asai2009), and MAFFT with the X-INS-i algorithm using MXSCARNA pairwise structural alignments and Contrafold base-pairing probabilities (Katoh & Toh Reference Katoh and Toh2008). The three structural alignments (for each gene region) were combined into a single alignment for each gene region using T-Coffee version 8.93 (Notredame et al. Reference Notredame, Higgins and Heringa2000). Subsequently, we filtered out ambiguously aligned regions as well as sites with a nucleotide in a single terminal and a gap in all other terminals. We defined ambiguous alignment as sites with a local consistency score (described by Notredame & Abergel Reference Notredame, Abergel and Andrade2003) less than 5. Scores from 5 to 9 (the highest) are, according to the documentation, considered to be correctly aligned with a probability exceeding 90%, given the underlying separate alignments. In other words, we kept alignment for which the three structural aligners generally agreed and excluded the rest.
Alignment of mrSSU
We downloaded the structural euascomycete mitochondrial 16S rRNA gene reference alignment from the Comparative RNA Web Site (http://www.rna.ccbb.utexas.edu; Cannone et al. Reference Cannone, Subramanian, Schnare, Collett, D'Souza, Du, Feng, Lin, Madabusi and Muller2002). We added our unaligned sequences to this profile using the L-INS-i algorithm of MAFFT and subsequently removed the profile and resulting gap-only columns. Ambiguously aligned sites were removed using Aliscore version 1.0 (Misof & Misof Reference Misof and Misof2009). All possible pairs of taxa were used to infer the consensus profile. The window size was set to 4 and gaps were treated as ambiguities.
Alignment of RPB1
Initial alignment was performed using the L-INS-i algorithm of MAFFT. Introns were identified and excised in accordance with the GenBank records submitted by James et al. (Reference James, Kauff, Schoch, Matheny, Hofstetter, Cox, Celio, Gueidan, Fraker and Miądlikowska2006). Finally, we trimmed the alignment to start with the first complete codon after the first intron reported by James et al. (Reference James, Kauff, Schoch, Matheny, Hofstetter, Cox, Celio, Gueidan, Fraker and Miądlikowska2006). The end of the alignment was trimmed to end after a third codon position and to keep the amount of missing data in the final alignment position below 50%.
Selection of partitioning scheme
The data were tentatively partitioned into seven initial subsets: ITS1, 5.8S, ITS2, mrSSU, and RPB1 first, second, and third codon positions, respectively. These subsets were subsequently input into PartitionFinder version 1.0.1 (Lanfear et al. Reference Lanfear, Calcott, Ho and Guindon2012) for an exhaustive search for the best-fitting partitioning scheme. We used the Bayesian Information Criterion (BIC) to select among models and partitioning schemes. We only considered proportional models (“branchlengths=linked”) across subsets (Pupko et al. Reference Pupko, Huchon, Cao, Okada and Hasegawa2002). The BIC has been shown to more accurately identify the generating model than the commonly used Akaike Information Criterion (AIC), assuming that the true generating model is included in the set of candidate models (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012).
Model selection
Although PartitionFinder reports a selected model for each of the partitions suggested, we performed a more thorough model selection from among the GTR family of likelihood models, including rate heterogeneity across sites and a proportion of invariable sites, on each of the final five subsets suggested by PartitionFinder. Model selection was performed using the Perl script MrAIC version 1.4.4 (Nylander Reference Nylander2004) in combination with PhyML version 20110919 (Guindon et al. Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010). As before, the BIC, with alignment length taken as sample size, was used to select among models. We included the number of branches in the number of free model parameters but we did not add an extra parameter for the topology. We selected among a reduced set of models with one, two, or six substitution rate categories, that is the ones available in frequently used software such as MrBayes version 3 (Ronquist & Huelsenbeck Reference Ronquist and Huelsenbeck2003). We consistently used six discrete gamma categories for modelling rate heterogeneity across sites. We modified MrAIC to improve PhyML search intensity by performing both NNI and SPR branch swapping and choosing the best outcome (the default is to perform only NNI branch swapping).
Model adequacy assessment
We assessed model adequacy (Goldman Reference Goldman1993; Bollback Reference Bollback2002), that is, the adequacy of the model selected to generate patterns similar to the sequence data observed. Model adequacy was assessed with PhyloBayes, using posterior predictive simulation from the GTR+Γ and F81+Γ+CAT models for each of the five subsets in the phylogenetic analysis. Simulations were performed across a random subset of 1000 trees drawn from the posterior distribution. We used the mean number of states per site ('site diversity') as the test statistic. Reported posterior predictive probabilities correspond to the fraction of times that the value from the posterior simulation exceeded the value observed from the data. Note that these are not probabilities in the classical sense, but rather describe the position of the test statistic derived from the observed data relative to the simulated data. The match to the model is perfect when the observed data fall in the centre of the simulated data, that is, when P is close to 0·5. Both extremely high and extremely low values of P signal poor adequacy of the model to reproduce the observed data. We considered P=<0·025 or P=>0·975 (i.e. the extreme 5%) as a significant departure from the model. We deliberately avoided the unconstrained (multinomial) likelihood as the test statistic (e.g. Bollback Reference Bollback2002) as all current implementations, unlike site diversity, require that all sites with gaps be excluded.
Phylogenetic analyses
PhyloBayes version 3.3b (Lartillot et al. Reference Lartillot, Lepage and Blanquart2009) was used to infer phylogeny under a baseline GTR+Γ model as well as under a F81+Γ+CAT and GTR+Γ+CAT model, using data from each of the five subsets separately as well as the concatenated data. Gamma distributed rate heterogeneity across sites was approximated as six discrete categories in all cases. Note that PhyloBayes does not implement a proportion of invariable sites. For concatenated data, we explored models with and without proportional branch lengths across subsets suggested by PartitionFinder. Under the CAT model (Lartillot & Philippe Reference Lartillot and Philippe2004), substitution rates are constant across sites and trees, whereas state frequencies are treated as a Dirichlet process with an infinite number of mixtures across sites, unobserved states at each site being united into a single state (Lartillot et al. Reference Lartillot, Brinkmann and Philippe2007). We used default priors, except that the prior on branch lengths was set to an exponential with a mean seeded by an exponential hyperprior with mean 0·1. We chose an exponential prior because empirical data suggest that true branch lengths are often exponentially distributed (Venditti et al. Reference Venditti, Meade and Pagel2010). Single-subset analyses were performed with three parallel runs, which were set to terminate automatically when the effective sample size of all model parameters exceeded 100 and the maximum discrepancy between runs of the likelihood and all diagnosed parameters descended below 0·1, discrepancy being measured as twice the difference in mean divided by the sum of standard deviations. The burn-in was set to a fifth of the chain length and is fixed by the software. In the end, however, we accepted only runs as converged if, in addition, the discrepancy of all parameters in the second half of the run was below 0·3. Concatenated analyses were performed in a similar manner, except that the three runs, for reasons of computational time, were treated as separate processes for a fixed number of cycles, 60 000. We subsequently applied the same convergence criteria as in the analyses of the individual partitions, except that we discarded the first half of the runs as burn-in and used every 10th tree from the second half of the runs to calculate a majority-rule consensus tree.
We also used MrBayes version 3.2.1 (Ronquist & Huelsenbeck Reference Ronquist and Huelsenbeck2003; Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) to infer phylogenies under a model with five partitions, each subset with the model favoured by MrAIC. Prior distributions included treating all tree topologies as equally likely, and (when applicable) a uniform (0·001, 200) distribution for the gamma shape parameter, a uniform (0, 1) distribution for the proportion of invariable sites, a (1, 1, 1, 1, 1, 1) Dirichlet for the rate matrix, independent beta (1, 1) distributions for the transition and transversion rates, and a (1, 1, 1, 1) Dirichlet for the state frequencies. The number of discrete categories used to approximate the gamma distribution was set to six in all analyses. We assumed an exponentially distributed branch length prior. The exponential distribution was parameterized with an empirical Bayes' approach (Ekman & Blaalid Reference Ekman and Blaalid2011), whereby the inverted branch length average calculated from a phylogeny generated with PhyML 3.0 online (Guindon et al. Reference Guindon, Lethiec, Duroux and Gascuel2005, Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010) was used as the exponential distribution rate parameter (d). This phylogeny was generated with a heuristic search involving NNI and SPR branch swapping from 10 random and one BIONJ tree under a GTR+I+Γ model. Three parallel Markov chain Monte Carlo (MCMC) runs were performed, each with four parallel chains and the temperature increment parameter set to 0·10 (Altekar et al. Reference Altekar, Dwarkadas, Huelsenbeck and Ronquist2004). The appropriate degree of heating was determined by observing swap rates between chains in preliminary runs. Every 1000th tree was sampled. Analyses were diagnosed for convergence every 106 generations in the last 50% of the tree sample and automatically halted when convergence was reached. Convergence was defined as an average standard deviation of splits (with frequency 0·1) between runs below 0·01. Finally, the potential scale reduction factor (PSRF) was monitored manually, and we only accepted runs with PSRF values smaller than 1·1 for all model parameters and all bipartitions.
Incongruence between the three genes (not the five partitions) was assessed by identifying conflicts between majority-rule consensus trees obtained by 1) maximum likelihood (ML) bootstrap analyses with PhyML 3.0 online and 2) Bayesian MCMC using PhyloBayes under a F81+Γ+CAT model. Each bootstrap analysis included 1000 bootstrap replicates and was performed under a GTR+I+Γ model. PhyloBayes analyses were performed in the same way as other analyses with the software described above. Majority-rule consensus trees were subsequently passed to Compat.py (Kauff & Lutzoni Reference Kauff and Lutzoni2002) for identification of conflicts. Tests were performed between all three pairs of genes. The cut-off for conflict identification was set to 0·7 in the ML analysis and 0·95 in the Bayesian analysis.
Branch attachment frequencies were calculated for selected taxa using Phyutility version 2.2.5 (Smith & Dunn Reference Smith and Dunn2008).
Marginal likelihoods of the data were calculated with Tracer version 1.5 (Rambaut & Drummond Reference Rambaut and Drummond2009) using importance sampling as suggested by Newton & Raftery (Reference Newton and Raftery1994) and modified by Suchard et al. (Reference Suchard, Kitchen, Sinsheimer and Weiss2003).
Results
Resources
The concatenated data, individual gene data used for assessing congruence, as well as all majority-rule consensus trees estimated from these data (including branch lengths and support values) are permanently filed in the TreeBASE repository (http://www.treebase.org) under study number 14978.
Partitioning and model selection
The selection of a partitioning scheme using PartitionFinder on the concatenated data indicated a preference for five subsets, viz. ITS1+ITS2, 5.8S, mrSSU, RPB1 first and second codon positions, and RPB1 third codon positions. The following models were selected by MrAIC under the Bayesian Information Criterion: HKY+Γ for the ITS1+ITS2, K80+I+Γ for the 5.8S, HKY+I+Γ for the mrSSU, GTR+Γ for the RPB1 1st+2nd positions, and HKY+I+Γ for the RPB1 3rd positions. Descriptive statistics for the five subsets, as well as the concatenated data, are found in Supplementary (see Supplementary Material , available online).
Gene tree incongruence
We identified two conflicts between gene trees. The very different placement of Degelia plumbea caused a deep conflict between the ITS on the one hand, and the mrSSU and RPB1 trees on the other hand in the ML bootstrap consensus but not in the Bayesian consensus. However, branch attachment frequencies reveal that in the Bayesian posterior tree sample obtained from ITS data, the three samples of D. plumbea cluster together with 100% posterior probability, and as sister group to Staurolemma omphalarioides with 98% posterior probability, a relationship that does not at all make sense from a morphological perspective. In the mrSSU and RPB1 trees, D. plumbea clusters, as expected from morphology, with D. atlantica and D. cyanoloma. Because of this deep incongruence, we excluded three specimens of D. plumbea (1–111) represented by ITS data. The second conflict, supported by both the ML and Bayesian consensus trees, occurred between the ITS and RPB1 and concerned the branching order among five closely related species of Pannaria. We did not exclude any taxa on account of this shallow incongruence.
Model adequacy
A GTR+Γ model was deemed significantly inadequate (P=1·000) in the case of the mrSSU and the RPB1 third codon positions, with poor performance also in the subsets consisting of ITS1 and ITS2 (P=0·970), 5.8S (P=0·844), and the RPB1 first and second codon positions (P=0·943). The F81+Γ+CAT model was not rejected for any of the five subsets (0·118≤P≤0·711).
Phylogeny from concatenated data
The ln marginal likelihoods calculated from the posterior samples produced by MrBayes (under a partitioned model, each subset with model selected by MrAIC) and PhyloBayes (under a F81+Γ+CAT model) were −19754·560 and −18454·879, respectively. The superiority of the F81+Γ+CAT model in this case, despite its very simple underlying substitution rate model, is not caused by differences in priors or the MCMC machinery across software, as analyses of each of the five subsets with MrBayes and PhyloBayes under a single GTR+Γ model produce closely matching marginal likelihoods (results not shown). The median posterior number of nucleotide frequency categories (‘profiles’) in the CAT model was 42. Apparently, there are substantial differences in nucleotide frequencies across our sequence data, leading to vastly different local instantaneous rates of substitution. We take the results from the F81+Γ+CAT model as our phylogenetic estimate, because this model clearly outperforms standard GTR family models with respect to model adequacy and likelihood. A majority-rule consensus tree with all compatible groups obtained with PhyloBayes under a F81+Γ+CAT model without subset-specific rate multipliers is shown in Fig. 1. Convergence statistics for this analysis translated to MrBayes standards (by feeding reformatted tree samples to ‘sumt’ of MrBayes) correspond to an average standard deviation of splits of 0·004 and a maximum topology PSRF of 1·003. We experienced severe convergence issues under the GTR+Γ+CAT (with and without subset-specific rate multipliers), as well as the F81+Γ+CAT with subset-specific rate multipliers despite very long runs, leading us to discard the results from these analyses.
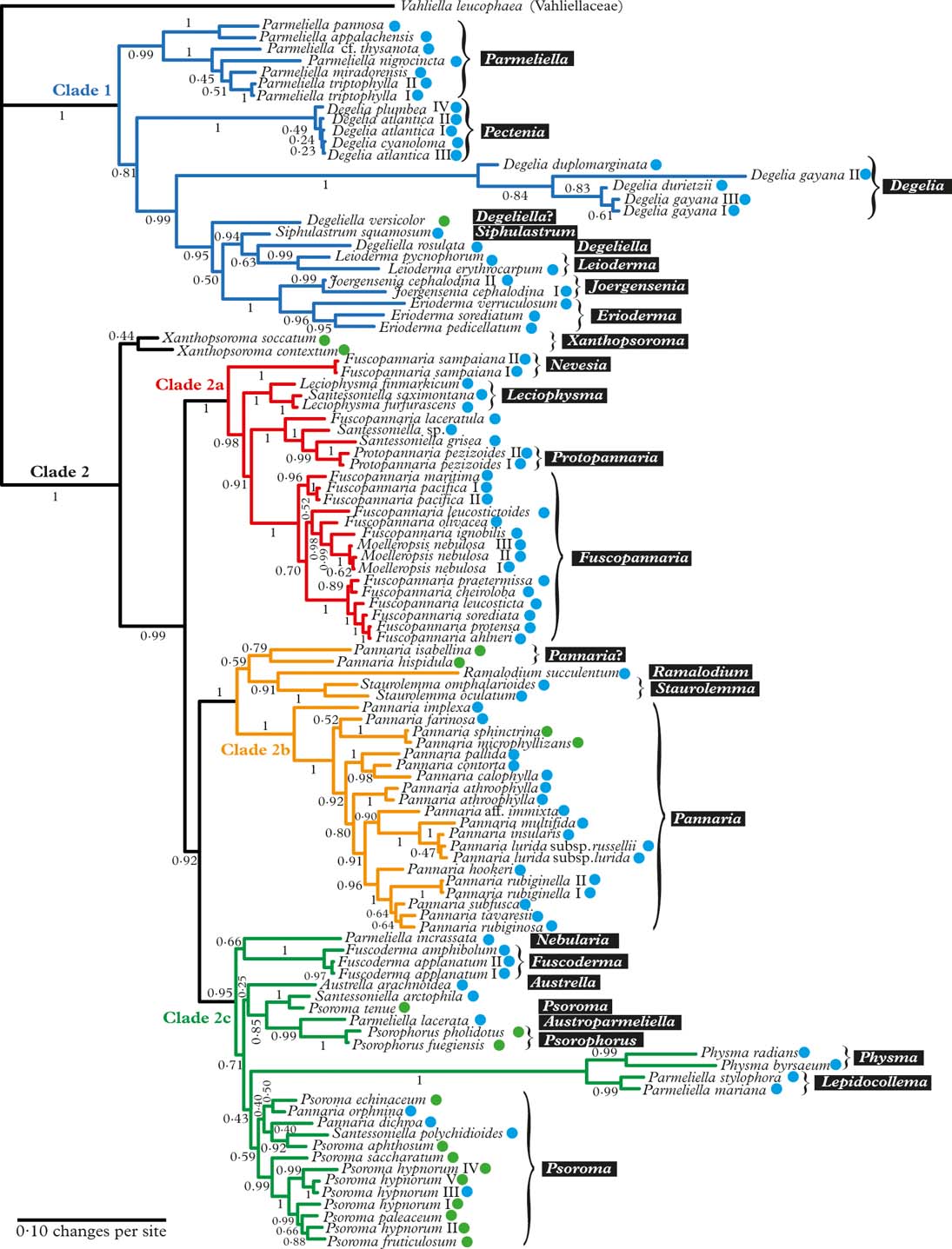
Fig. 1. Majority-rule consensus tree with all compatible groups and average branch lengths resulting from Bayesian MCMC with PhyloBayes under an F81+Γ+CAT model on concatenated data from ITS, mrSSU, and RPB1. Bayesian posterior probabilities are indicated. Vahliella leucophaea in the Vahliellaceae is the outgroup. Names generally follow Jørgensen (Reference Jørgensen2003), although our interpretation of generic affinities (resulting from this phylogeny, other published phylogenies cited in the text, as well as morphological data) is indicated with white text against a black background. Four main clades (1, 2a, 2b, and 2c) are indicated in colour. Roman numerals are used to distinguish specimens of the same taxon. Coloured dots after taxon names indicate the type of primary photobiont (blue=cyanobacterial, green=green-algal photobiont). In colour online.
Discussion
Model adequacy, robustness, and branch support
Spuriously high branch support in Bayesian phylogenetics as sometimes reported (summarized by Alfaro & Holder Reference Alfaro and Holder2006) can have two explanations, disregarding MCMC machinery failure: mis-specified priors and/or under-parameterized models (Yang Reference Yang2006: 178–179). We safeguarded against the bias from a mis-specified prior on branch lengths by use of a hyperprior (in PhyloBayes) or an empirical Bayes prior (in MrBayes) (Kolaczkowski & Thornton Reference Kolaczkowski and Thornton2007; Ekman & Blaalid Reference Ekman and Blaalid2011). Bayesian branch support estimates seem to be particularly sensitive to model under-parameterization (Buckley Reference Buckley2002; Huelsenbeck & Rannala Reference Huelsenbeck and Rannala2004; Lemmon & Moriarty Reference Lemmon and Moriarty2004; Brown & Lemmon Reference Brown and Lemmon2007). Therefore, we conducted an assessment of model adequacy in an attempt to identify a model that was capable of reproducing patterns of the observed data. We found that ordinary GTR family models, including rate heterogeneity across sites, were inadequate as long as spatial heterogeneity in nucleotide frequency, and consequently local differences in the instantaneous rates of substitution, were not included in the model. A model incorporating this process, in this case CAT (Lartillot & Philippe Reference Lartillot and Philippe2004), was found to be adequate for all our data subsets as measured by nucleotide site diversity. Branch support generated from an adequate model is unlikely to be overestimated. Indeed, average support for internal branches in the consensus tree estimated by MrBayes (not shown here but included in the TreeBASE submission) was on average 2·1% higher than the corresponding tree obtained with PhyloBayes (87·7% vs. 85·6%), and three branches in the MrBayes consensus had distinctly higher support to the point where it would affect conclusions drawn from the analysis.
Gene tree conflicts
The ML phylogeny based on the ITS data conflicted with the corresponding mrSSU and RPB1 ML phylogenies regarding the position of Degelia plumbea, which is represented in the ITS tree by three different samples, all from western Norway. During the course of this investigation, identical ITS sequences were recovered from several more specimens, also from Norway, that are not reported here. The lack of apparent conflict regarding the position of D. plumbea between the Bayesian gene consensus trees is ostensibly caused by poor backbone support in the ITS consensus tree. The poor support is not caused by rogue behaviour of D. plumbea, as branch attachment frequencies indicate that D. plumbea clusters on a long branch as sister group to Staurolemma with 98% posterior probability. This association cannot be reconciled with morphology. In the mrSSU and RPB1 Bayesian as well as ML phylogenies, D. plumbea clusters, as expected from morphology, with D. atlantica and D. cyanoloma.
The ITS sequences we have recovered from Degelia plumbea may ultimately prove to be non-orthologous. The same potential non-orthologue was captured by Ekman & Jørgensen (Reference Ekman and Jørgensen2002) and fell outside the Pannariaceae in their phylogeny. Interestingly, what seems to be the orthologue was recently reported by Otálora et al. (Reference Otálora, Salvador, Martínez and Aragón2013), who used different PCR primers and sampled from a different geographical area, southern and central Spain. There are, however, no reported cases of ascomycetes containing a non-orthologous rDNA sequence that was transformed extensively by processes not handled well by current phylogenetic likelihood models. We do not claim the ITS sequences observed in D. plumbea to be the first such case, because crucial experimental evidence of intragenomic variation is still lacking. However, our observations call for further scrutiny.
A second gene tree conflict involved the branching order between Pannaria rubiginosa, P. rubiginella, P. tavaresii, P. subfusca, and P. hookeri in the ITS and RPB1 trees. These taxa form a group of closely related species (Jørgensen Reference Jørgensen1978). Shallow conflicts like these may represent incomplete lineage sorting (a.k.a. deep coalescence). In such instances, concatenation of data from several genes has been shown to be a poor method for estimating the species tree (Edwards et al. Reference Edwards, Liu and Pearl2007; Kubatko & Degnan Reference Kubatko and Degnan2007). Unlike in the case of Degelia plumbea, pointing out a single culprit offending congruence is not possible. We did not proceed to exclude any data from the concatenated analysis, as we were primarily interested in inferring boundaries and relationships at the genus level. We note, however, that inferred relationships from the concatenated data between taxa involved in this conflict must be interpreted with caution.
Overview of the Pannariaceae
The Pannariaceae, as currently circumscribed, has previously been shown to be monophyletic (Wedin & Wiklund Reference Wedin and Wiklund2004; Wedin et al. Reference Wedin, Jørgensen and Wiklund2007, Reference Wedin, Wiklund, Jørgensen and Ekman2009; Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011; Spribille & Muggia Reference Spribille and Muggia2013), and falls into two major clades (Clade 1 and 2 in Fig. 1); to some extent these coincide with the formation of a secondarily developed margin of thalline origin in the apothecia of the second clade and the corresponding absence of such a margin in the first clade. There are several exceptions to this rule, however, Joergensenia having a well-developed secondary thalline margin, as well as species scattered in the second clade lacking a thalline margin, mainly in gelatinous taxa with a cyanobacterial photobiont. Clade 1 includes Parmeliella, Degelia, Degeliella, Siphulastrum, Joergensenia, Leioderma, and Erioderma. According to Muggia et al. (Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011), the genus Leptogidium, not included in our study, also belongs here. Clade 2 consists of three subclades (2a, b, c) and Xanthopsoroma. Clade 2a includes Fuscopannaria s. lat. (incl. Moelleropsis), Leciophysma, Protopannaria and some species referred to Santessoniella. The recently described Steineropsis (Spribille et al. Reference Spribille, Pérez-Ortega, Tønsberg and Schirokauer2010), although not included in our study, also belongs here (Spribille & Muggia Reference Spribille and Muggia2013). Clade 2b contains Pannaria, Ramalodium, and Staurolemma, and Clade 2c includes Psoroma s. lat., Fuscoderma, Austrella, Santessoniella , Psorophorus, and Physma. The genus Xanthopsoroma falls outside these clades in our phylogeny. Support for its monophyly is very weak, but support for branches on either side of the genus is high, indicating that Xanthopsoroma, as currently understood, is either monophyletic or a paraphyletic grade.
Generic taxonomy and biogeography
We recognize 30 genera altogether in the Pannariaceae, although some are provisional. The two largest genera, Pannaria and Lepidocollema, are mostly tropical with some extensions through the subtropical region into warm temperate regions. The highest number of genera is found in the Southern Hemisphere, particularly in South America, possibly reflecting a long and complex biogeographical history in that part of the world. Three genera are confined to the Northern Hemisphere, two in the Atlantic-Mediterranean part of Europe (Nevesia and Pectenia) and one in North America (Fuscopannaria). Fuscopannaria is the largest genus of the family in the temperate zone and is particularly species-rich in the North Pacific region, although a few species extend into the Southern Hemisphere. Psoroma s. str. is genuinely bipolar, with far more species in the Southern Hemisphere than elsewhere.
Synopsis of Genera in the Pannariaceae
In this section, we briefly treat all genera currently accepted by us, the delimitation of which mostly emerge from the phylogenetic estimate (Fig. 1), but also on grounds of previous phylogenetic estimates as well as morphological and chemical data. We also include genera that were not part of the phylogeny, which we refer to the family based on other than phylogenetic evidence. Finally, we present an identification key to the accepted genera.
Genera in bold font (whether italicized or not) are accepted genera. A star in front of the name indicates that no member of the genus was included in our phylogeny. Genera in regular font are names for genera that are considered here as synonyms and should be abandoned. We provide full descriptions of newly established genera.
Austrella P. M. Jørg. (Fig. 5D) was described by Jørgensen (Reference Jørgensen2004) for the type species A. arachnoidea and A. brunnea, which are characterized by the formation of apothecia from non-lichenized fungal hyphae, a thick subhymenium of densely packed tissue, and the lack of an apical apparatus in the asci. We provisionally retain the genus as originally conceived, although we note that Austrella has an uncertain position within Clade 2c.
Austroparmeliella (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB807983
Parmeliella sect. Austroparmeliella P. M. Jørg., Bibl. Lich. 88: 244 (2004).
Generitype: A. lacerata (P. M. Jørg.) P. M. Jørg. (Fig. 5C)
Thallus bluish grey, composed of squamules that form a lace-like crust. Squamules usually deeply incised, 2–3 mm wide, up to 75 µm thick; upper cortex 10–15 µm thick, cellular; medulla up to 50 µm thick, of loosely arranged, intricate hyphae enclosing clusters of Nostoc; lower cortex of a single cell-layer or lacking in parts of the thallus.
Apothecia frequent, often grouped, c. 1 mm diam., becoming convex at maturity, with red-brown disc surrounded by pale rim; proper exciple paraplectenchymatous, 30–50 µm wide. Subhymenium colourless, flat, 100–150 µm thick, of intricately interwoven hyphae. Hymenium 100–150 µm µm high, I+ deep blue. Asci cylindrical, with apical amyloid ring structure, 8-spored; ascospores colourless with smooth wall, broadly ellipsoid, non-septate.
Pycnidia not observed.
Chemistry
No lichen substances found (Jørgensen Reference Jørgensen2004).
Notes
This is a genus of small, Southern Hemispheric Parmeliella-like species with finely divided squamules, often with cortex on all sides. A further difference from Parmeliella s. str. is the narrow, flat, colourless subhymenium, as opposed to the often lentil-shaped, brownish subhymenium in Parmeliella. Our phylogeny suggests a sister-group relationship with Psorophorus, the members of which differ in the hemiamyloid hymenia and in forming thalline apothecial margins. Five species of Austroparmeliella are recognized here, the four species treated by Jørgensen (Reference Jørgensen2004) and ‘Santessoniella’ elongata (Henssen Reference Henssen1997). The latter, although not known to produce apothecia, is transferred here to Austroparmeliella on account of the presence of a lower cortex.
Degelia Arv. & D. J. Galloway (Fig. 2A) was originally described to accommodate Coccocarpia-like, Southern Hemisphere species with apothecia similar to Parmeliella (Arvidsson & Galloway Reference Arvidsson and Galloway1981), but with different asci (without an apical amyloid tube). Jørgensen & James (Reference Jørgensen and James1990) added the three species of the Northern Hemispheric ‘Parmeliella’ plumbea group known at the time (D. plumbea, D. atlantica, and D. ligulata), and later Blom & Lindblom (Reference Blom and Lindblom2009) added one more species, Degelia cyanoloma. A separate section, Amphiloma P. M. Jørg. & P. James, with D. plumbea as its type species, was established for this group of species (Jørgensen & James Reference Jørgensen and James1990). Members of sect. Amphiloma possess a Nostoc photobiont, whereas other species are lichenized with Scytonema. A third section, Frigidae P. M. Jørg., was described by Jørgensen (Reference Jørgensen2004) for three subantarctic species with a thick paraplectenchymatous upper cortex and a poorly developed secondary thalline corona. The type species of this section is D. subcincinnata (Nyl.) P. M. Jørg.
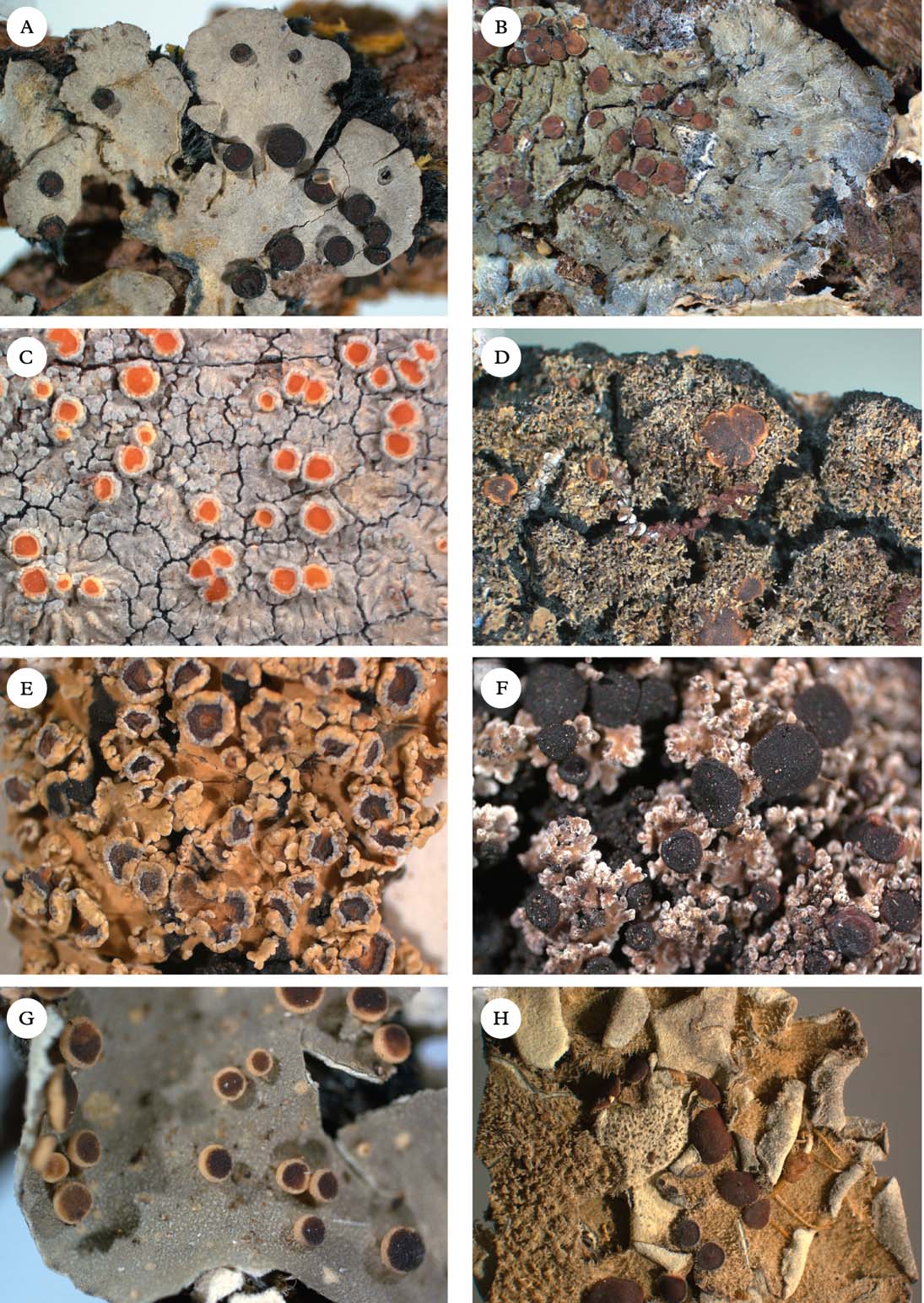
Fig. 2. Representatives of Clade 1. A, Degelia gayana; B, Pectenia plumbea; C, Degeliella rosulata; D, Parmeliella triptophylla; E, Joergensenia cephalodina; F, Siphulastrum squamosum; G, Leioderma pycnophorum; H, Erioderma leylandii. Photographs: Jan Berge. In colour online.
Our phylogeny (Fig. 1) indicates that Degelia as currently understood is non-monophyletic and that the monophyletic section Amphiloma should be recognized as a separate genus. Therefore, we introduce the new name Pectenia for this section (see below).
Degelia sect. Frigidae was not represented in our phylogeny. However, a member of this section, D. symptychia (Tuck.) P. M. Jørg., was represented in the phylogeny of Spribille & Muggia (Reference Spribille and Muggia2013) and was shown to belong in Steinera in the Koerberiaceae. Unfortunately, sequence data is currently lacking for the type species, D. subcincinnata, which is why we refrain from further taxonomic and nomenclatural changes at the moment.
In Degelia s. str., there may be a problem with heterogeneity in what has been treated as D. gayana, the type species, unless this species-level non-monophyly is caused by incomplete lineage sorting or another (undetected) case of non-orthology (Fig. 1).
Degeliella P. M. Jørg. (Fig. 2C) was described by Jørgensen (Reference Jørgensen2004) to accommodate D. rosulata (P. M. Jørg. & D. J. Galloway) P. M. Jørg., the type species, and D. versicolor (Müll. Arg.) P. M. Jørg. (Jørgensen Reference Jørgensen2004). Morphologically, it was separated from Degelia on account of the non-amyloid hymenium and ascus, a feature shared by the closely related genera Siphulastrum and Leioderma (Galloway & Jørgensen Reference Galloway and Jørgensen1987; Jørgensen Reference Jørgensen1998). Degeliella rosulata possesses a cyanobacterial photobiont and smooth ascospores, whereas D. versicolor has a green-algal primary photobiont and warted ascospores. In our phylogeny, the type species D. rosulata forms a monophyletic group with fair support (0·94 posterior probability), together with Siphulastrum and Leioderma. Degeliella versicolor (Fig. 7E) is unlikely to be monophyletic together with the type species and may deserve generic recognition (see Psoromaria).
Erioderma Fée (Fig. 2H) includes more than 30 species. The genus has a complex chemistry (Jørgensen & Arvidsson Reference Jørgensen and Arvidsson2002) and is recognized by an ascomatal ontogeny unique to the family (Keuck Reference Keuck1977).
Fuscoderma (D. J. Galloway & P. M. Jørg.) P. M. Jørg. & D. J. Galloway (Fig. 5B) is a genus of five known species, two of which are represented in our phylogeny. They form a monophyletic group and are obviously distantly related to Leioderma, under which they were originally placed as a subgenus (Galloway & Jørgensen Reference Galloway and Jørgensen1987). Fuscoderma belongs in Clade 2c, where it is the sister of the Andean genus Nebularia (see below). Fuscoderma is recognized by squamulose to subfoliose, heteromerous thalli with a Nostoc photobiont and brownish tomentum on the lower side, a non-amyloid hymenium (except the gel surrounding asci), a lack of amyloid apical structures in the asci, and the production of vicanicin and/or norvicanicin (Jørgensen & Galloway Reference Jørgensen and Galloway1989).
Fuscopannaria P. M. Jørg. (Fig. 3D) is a genus of c. 50 species that was separated from Pannaria on account of the hemiamyloid hymenium, asci with an amyloid apical ring structure, and the production of fatty acids and terpenoids but not pannarin (Jørgensen Reference Jørgensen1978, Reference Jørgensen1994). In addition, most species are small-squamulose and form apothecia with a variably developed thalline margin, which can sometimes even be missing.

Fig. 3. Representatives of Clade 2a. A, Nevesia sampaiana; B, Protopannaria pezizoides; C, Leciophysma finmarkicum; D, Fuscopannaria leucosticta. Photographs: Jan Berge. In colour online.
The majority of the species, including the type F. leucosticta (Tuck.) P. M. Jørg., form a monophyletic group if F. sampaiana and F. laceratula are excluded. However, whereas F. sampaiana is included here in the newly described genus Nevesia (see below), we refrain from a formal placement of F. laceratula, awaiting improved taxon sampling. ‘Fuscopannaria’ laceratula is set apart by its combination of secondary chemistry (atranorin) and a Scytonema-like photobiont (Jørgensen Reference Jørgensen2005a ).
Moelleropsis nebulosa (Hoffm.) Gyeln. (Fig. 6A) is nested within Fuscopannaria, as suggested already by Ekman & Jørgensen (Reference Ekman and Jørgensen2002), although scarce taxon sampling prevented them from definitively placing Moelleropsis in synonymy. This situation has unfortunate nomenclatural consequences, since Moelleropsis is an older name than Fuscopannaria. We retain the use of Fuscopannaria, including Moelleropsis, pending a final decision based on a proposal to conserve Fuscopannaria against Moelleropsis (Jørgensen et al. Reference Jørgensen, Ekman and Wedin2013).
Subgenus Micropannaria P. M. Jørg. was established to comprise F. leucophaea and related species (Jørgensen Reference Jørgensen1994) but was later described as a separate genus, Vahliella P. M. Jørg. (Jørgensen Reference Jørgensen2008); it is now placed in the currently monogeneric Vahliellaceae (Wedin et al. Reference Wedin, Jørgensen and Ekman2011; Spribille & Muggia Reference Spribille and Muggia2013).
*Homothecium A. Massal. is a genus of five small-sized species with a gelatinous thallus from southern South America. The genus is morphologically and anatomically similar to Ramalodium, from which it differs mainly in the annular exciple (cupular in Ramalodium) and presence of an apical ring structure in the ascus (none in Ramalodium) (Henssen Reference Henssen1965, Reference Henssen1979). Although currently referred to the Collemataceae (Lumbsch & Huhndorf Reference Lumbsch and Huhndorf2010) and not included in our phylogeny, we provisionally treat Homothecium as another genus in the Pannariaceae with non-septate ascospores and a gelatinous thallus.
Joergensenia Passo et al. (Reference Passo, Stenroos and Calvelo2008) (Fig 2E) and appears in our phylogeny as the sister group to the morphologically and chemically very different Erioderma. Joergensenia (Fig. 1) is aberrant in being the only genus in Clade 1 with a secondarily developed thalline margin in the apothecia (i.e. not an ontogenetically ‘true proper margin’). The thalline “corona” in the apothecia of a few species of Degelia and Degeliella is, according to Henssen & James (Reference Henssen and James1980), not an ordinary thalline margin. Furthermore, Joergensenia is characterized by its strongly amyloid cap-shaped plug in the ascus apex.
*Kroswia P. M. Jørg. is a small genus (Jørgensen Reference Jørgensen2002) of three paleotropical species (Jørgensen & Gjerde Reference Jørgensen and Gjerde2012) that were formerly believed to be closely related to Physma (Swinscow & Krog Reference Swinscow and Krog1988). However, the discovery of fertile material revealed characters in the hymenium, suggesting a close relationship with Fuscopannaria (Jørgensen Reference Jørgensen2007). The globose, brown-pigmented ascospores are unique in the family.
Leciophysma Th. Fr. (Fig. 3C) was treated in detail by Henssen (Reference Henssen1965). The genus is monophyletic if Santessoniella saximontana P. M. Jørg. & T. Sprib. is included. Leciophysma is distantly related to the type species of Santessoniella, S. polychidioides, which is morphologically similar and sometimes difficult to distinguish from Leciophysma.
Leioderma Nyl. (Fig 2G) forms a monophyletic group in a clade together with Degeliella, Siphulastrum, Joergensenia, and Erioderma. Morphologically, Leioderma is similar to Erioderma, from which it differs by the lack of thallus chemistry. Leioderma as circumscribed here corresponds to Leioderma subgenus Leioderma of Galloway & Jørgensen (Reference Galloway and Jørgensen1987), whereas subgenus Fuscoderma corresponds to the genus Fuscoderma (see above).
*Leightoniella Henssen, with its only known species L. zeylanica (Cromb. ex Leight.) Henssen, is known only from the type material, which was described in detail by Henssen (Reference Henssen1965). This genus has so far been classified in the Collemataceae (e.g., Lumbsch & Huhndorf Reference Lumbsch and Huhndorf2010) and is characterized by the periclinally arranged hyphae in the exciple and the production of ‘supporting tissue’ along the thalline margin and thallus stalk (Henssen Reference Henssen1965). The thallus is gelatinous with cyanobacteria and ascospores are simple. Although not included in our phylogeny, we provisionally treat Leightoniella as another member of the Pannariaceae with a gelatinous thallus and simple ascospores.
Lepidocollema Vain. was described by Vainio (Reference Vainio1890) to accommodate a single gelatinous, homoiomerous Parmeliella-like species with a Nostoc photobiont, L. carassense Vain., which has been collected only once, in Brazil. Vainio also noted the striking similarity with the apothecia of Parmeliella mariana (as Pannaria mariana), although he acknowledged the difference in thallus anatomy, P. mariana being heteromerous (albeit also contaning Nostoc). Although material of the type species was unavailable to us, we accept the genus here for a total of 24 tropical species, including ‘Parmeliella’ stylophora and ‘P.’ mariana (Figs 1 & 5E). Lepidocollema as understood here is characterized by the formation of large, flat rosettes on a thick layer of rhizohyphae, the presence of a cellular thalline cortex, apothecia with a thalline margin, asci with a wide apical ring structure, and thin-walled ascospores. The thallus is heteromerous in all species except the type species. The genus is sister to Physma (for differences see that genus). Most of the species have been treated in Parmeliella (e.g. Jørgensen & Galloway Reference Jørgensen and Galloway1992), with which they are only distantly related.
* Leptogidium Nyl. was recently re-established for the type species L. dendriscum (Nyl.) Nyl., as well as L. contortum (Henssen) T. Sprib. & Muggia and L. stipitatum (Vězda & W. A. Weber) T. Sprib. & Muggia (Muggia et al. Reference Muggia, Nelson, Wheeler, Yakovchenko, Tønsberg and Spribille2011). These species have traditionally been treated in Polychidium (Henssen Reference Henssen1963), from which they are easily distinguished by the photobiont being Scytonema instead of Nostoc.
Moelleropsis Gyeln. (Fig. 7A), with its single species M. nebulosa (Hoffm.) Gyeln., is nested within Fuscopannaria and should be reduced into synonymy with that genus.
Nebularia P. M. Jørg. gen. nov.
MycoBank No.: MB807981
Fuscodermi similis, sed thallo subtus sine tomento fusco et hymenio in iodo toto coerulescenti.
Generitype: Nebularia incrassata (P. M. Jørg.) P. M. Jørg.
(Fig. 5A)
Thallus brownish, composed of up to 3 mm wide squamules with up to 0·25 mm wide, thickened, digitate lobes; upper cortex prominent, cellular, up to 70 µm thick; medulla c. 150 µm thick, of intricately interwoven hyphae enclosing often densely packed clusters of Nostoc, individual cells 5–7 µm diam.
Apothecia up to 1·5 mm diam., reddish brown, flat, with paler, prominent rim; proper exciple paraplectenchymatous, up to 80 µm wide. Subhymenium poorly delimited, colourless with loosely interwoven hyphae, containing photobiont cells that penetrate marginally from below. Hymenium up to 150 µm thick, I+ deep blue. Asci cylindrical, with distinct apical amyloid tube, 8-spored; ascospores colourless with rugulose wall, globose to ellipsoid, non-septate.
Pycnidia not observed.
Chemistry
No lichen substances found (Jørgensen Reference Jørgensen2000; Jørgensen & Palice Reference Jørgensen and Palice2010).
Etymology
From Latin nebula (fog) and –aris (belonging to), as the species grows in ‘selvas nubladas’ (=foggy forests).
Notes
Nebularia is an Andean genus comprised of only two species, the type species N. incrassata and N. psoromoides. Both species were originally referred to Parmeliella, with which they are only distantly related according to our phylogeny. In our phylogeny, Nebularia belongs in Clade 2c, although support for relationships within that clade is poor. Nebularia is morphologically similar to Fuscoderma in the shiny apothecia with a prominent apothecial rim, and in photobiont cells penetrating into the subhymenium. The latter character is unique to the two genera within the family. However, the amyloid, I+ deep blue hymenium, as well as the absence of tomentum on the lower surface, sets Nebularia apart from Fuscoderma, which has a hemiamyloid hymenium and brown tomentum on the lower surface.
Nevesia P. M. Jørg., L. Lindblom, Wedin & S. Ekman gen. nov.
MycoBank No.: MB807982
Thallus crustaceo-squamulosus hypothallo distincto positus, castaneus cum sorediis granulatis eburneis, sine acidis lichenis. Apothecia matura et pycnidia ignota.
Generitype: Nevesia sampaiana (Tav.) P. M. Jørg., L. Lindblom, Wedin & S. Ekman.
(Fig. 3A)
Thallus consisting of 2–3 mm wide, chestnut brown, appressed, up to 200 µm thick squamules; hypothallus well developed, blue-black; upper cortex cellular, 50–60 µm thick; algal layer 50–70 µm thick, Nostoc cells 6–8 µm diam., in clusters; medulla 40–80 µm thick, of intricate, 3–4 µm wide hyphae, forming a lax plectenchyma, gradually merging into the hypothallus.
Apothecia with thalline margin, extremely rare, known only in an immature state without developed asci. Hymenium hemiamyloid.
Pycnidia not known.
Chemistry
No lichen substances found (Jørgensen Reference Jørgensen1978).
Etymology
Named in honour of the Portuguese lichenologist Carlos das Neves Tavares (1914–1972), who first recognized N. sampaiana (as Pannaria sampaiana) at species level (Tavares Reference Tavares1950). He had a keen interest in, and substantial knowledge of, the Pannariaceae, which he generously shared with PMJ when he started working on this group.
Notes
Nevesia is a monospecific genus. Originally included in Pannaria, its only species was later transferred to Fuscopannaria (Jørgensen Reference Jørgensen1994). It is not known with mature apothecia, and its former classification was essentially based on overall morphology, secondary chemistry, and the observation of a hemiamyloid reaction of the hymenium in immature apothecia (Jørgensen Reference Jørgensen1978, Reference Jørgensen1994). It differs from most species of Fuscopannaria in having a very well-developed hypothallus, and in the chestnut-coloured thallus lacking lichen substances. In our phylogeny, Nevesia is sister to a large group containing mainly Leciophysma, Protopannaria, and Fuscopannaria.
Pannaria Del. (Fig. 4C) is a genus of c. 80 species, with Pannaria rubiginosa being the type species. The genus is recognized by a squamulose or foliose thallus, apothecia with a thalline margin, an amyloid hymenium, asci without internal amyloid apical structures, and the presence of pannarin and related substances (Jørgensen Reference Jørgensen1994, Reference Jørgensen2001a ). Historically, Pannaria included squamulose species containing a Nostoc photobiont and apothecia with a thalline margin.

Fig. 4. Representatives of Clade 2b. A, Ramalodium succulentum; B, Staurolemma omphalarioides; C, Pannaria rubiginosa. Photographs: Jan Berge. In colour online.
Most members of Pannaria included here form a monophyletic group, although a few may belong elsewhere, such as P. isabellina, P. hispidula, P. orphnina, and P. dichroa. Pannaria isabellina (Fig. 6A) and P. hispidula (Fig. 6B) form a poorly supported group with Staurolemma and Ramalodium. Together with Pannaria s. str., they form the strongly supported group we refer to here as Clade 2b. It is currently impossible to confirm or rule out the possibility that P. isabellina and P. hispidula belong in Pannaria. Also, Pannaria dichroa and P. orphnina appear to be currently misclassified and belong to Clade 2c (see discussion under Psoroma).
Our results support the notion that Pannaria also includes taxa with a green-algal photobiont (in our phylogeny represented by P. sphinctrina and P. microphyllizans), previously treated in Psoroma (Jørgensen Reference Jørgensen2001a ). There is no support for the recognition of subgenus Lepidoleptogium (A. L. Sm.) P. M. Jørg., as the type species L. montagnei A. L. Sm. is a member of the Pannaria immixta complex, which is nested inside Pannaria s. str. in our phylogeny.
Parmeliella Müll. Arg. (Fig. 2D) was originally established for squamulose members of the Pannariaceae with apothecia lacking a thalline margin. In later treatments (e.g., Jørgensen Reference Jørgensen1978), it was restricted to include species with an amyloid apical ring structure and a lack of lichen substances in the thallus. Even after the separation of Degelia (see above), Parmeliella remained heterogeneous. Most species of Parmeliella form a monophyletic group, although P. incrassata, P. lacerata, P. mariana, and P. stylophora are obviously misclassified. However, Parmeliella can be retained as a monophyletic entity, including the type species P. triptophylla and the majority of species in the genus, if the tropical Parmeliella mariana group is moved to Lepidocollema, and P. lacerata and P. incrassata are referred to the new genera Austroparmeliella and Nebularia, respectively. In its revised circumscription, Parmeliella is a mostly temperate genus including small-squamulose species, generally without chemical substances and apothecia without a thalline margin, but with an amyloid hymenium producing asci with an internal apical tube structure.
It is noteworthy that the likewise tropical Parmeliella pannosa (Sw.) Nyl., which is often confused with P. mariana, belongs in Parmeliella s. str. Parmeliella pannosa has a narrow and tube-like amyloid apical structure typical of the genus, whereas species of Lepidocollema have a broader ring-like apical structure.
Pectenia P. M. Jørg., L. Lindblom, Wedin & S. Ekman nom. et stat. nov.
MycoBank No.: MB807984
Degelia sect. Amphiloma (Fr.) P. M. Jørg. & P. James, Bibl. Lich. 38: 261 (1990).
Generitype: Pectenia plumbea (Lightf.) P. M. Jørg., L. Lindblom, Wedin & S. Ekman.
(Fig. 2B)
Thallus blue-grey, placodioid, appearing thick and rigid, in orbicular patches up to 10 cm diam., up to 250 µm thick. Upper cortex cellular, up to 40 µm thick. Photobiont layer 60–100 µm thick, with Nostoc cells 6–8 µm diam., in clusters. Medulla up to 150 µm thick, composed of parallel, branched, short-celled, horizontally aligned hyphae forming a compact plectenchyma, gradually merging into the hypothallus. Hypothallus thick, felt-like, blue-black, often extending beyond the ascending marginal lobes.
Apothecia laminal, usually abundant, biatorine with brown disc and a paler rim. Proper exciple up to 100 µm wide, consisting of isodiametric cells. Subhymenial layers pale yellowish brown, up to 150 µm thick, composed of intricately interwoven hyphae. Hymenium 100–150 µm high, colourless except for brown pigment in uppermost part, I+ persistently blue. Paraphyses unbranched. Asci clavate to cylindrical, with an apical dark-amyloid plug. Ascospores 8 per ascus, colourless, ellipsoid with smooth wall and without perispore, non-septate.
Pycnidia infrequent, mostly marginal, protruding, black, up to 0·2 mm wide. Conidiophores short-celled, producing conidia terminally and laterally. Conidia bacilliform, 1–3×1 µm.
Chemistry
No lichen substances found (Jørgensen & James Reference Jørgensen and James1990).
Etymology
From the Latin generic name of scallop, Pecten, due to the grooved scallop-like pattern often found on the upper surface of the species in this genus.
Notes
The name Amphiloma cannot be used at generic level, since it is occupied by two older homonyms (Jørgensen Reference Jørgensen1978). Consequently, we establish the new name Pectenia based on sect. Amphiloma and with the same type species, P. plumbea. Pectenia is mainly confined to Europe and adjacent Africa, mostly along the Atlantic coast. However, P. plumbea also occurs in a restricted region in North-East America (Blom & Lindblom Reference Blom and Lindblom2009; Richardson et al. Reference Richardson, Anderson, Cameron and Seaward2010).
Physma A. Massal. (Fig. 5F), previously treated in the Collemataceae, belongs in the Pannariaceae, as also shown by Wedin et al. (Reference Wedin, Wiklund, Jørgensen and Ekman2009) and Otálora et al. (Reference Otálora, Aragón, Molina, Martínez and Lutzoni2010). In our phylogeny, Physma is the sister group to the Parmeliella mariana group, referred here to Lepidocollema. Physma is characterized by a leathery thallus with a dense upper pseudocortex (unlike the cellular cortex in Lepidocollema), and thick-walled ascospores with a markedly swollen epispore.
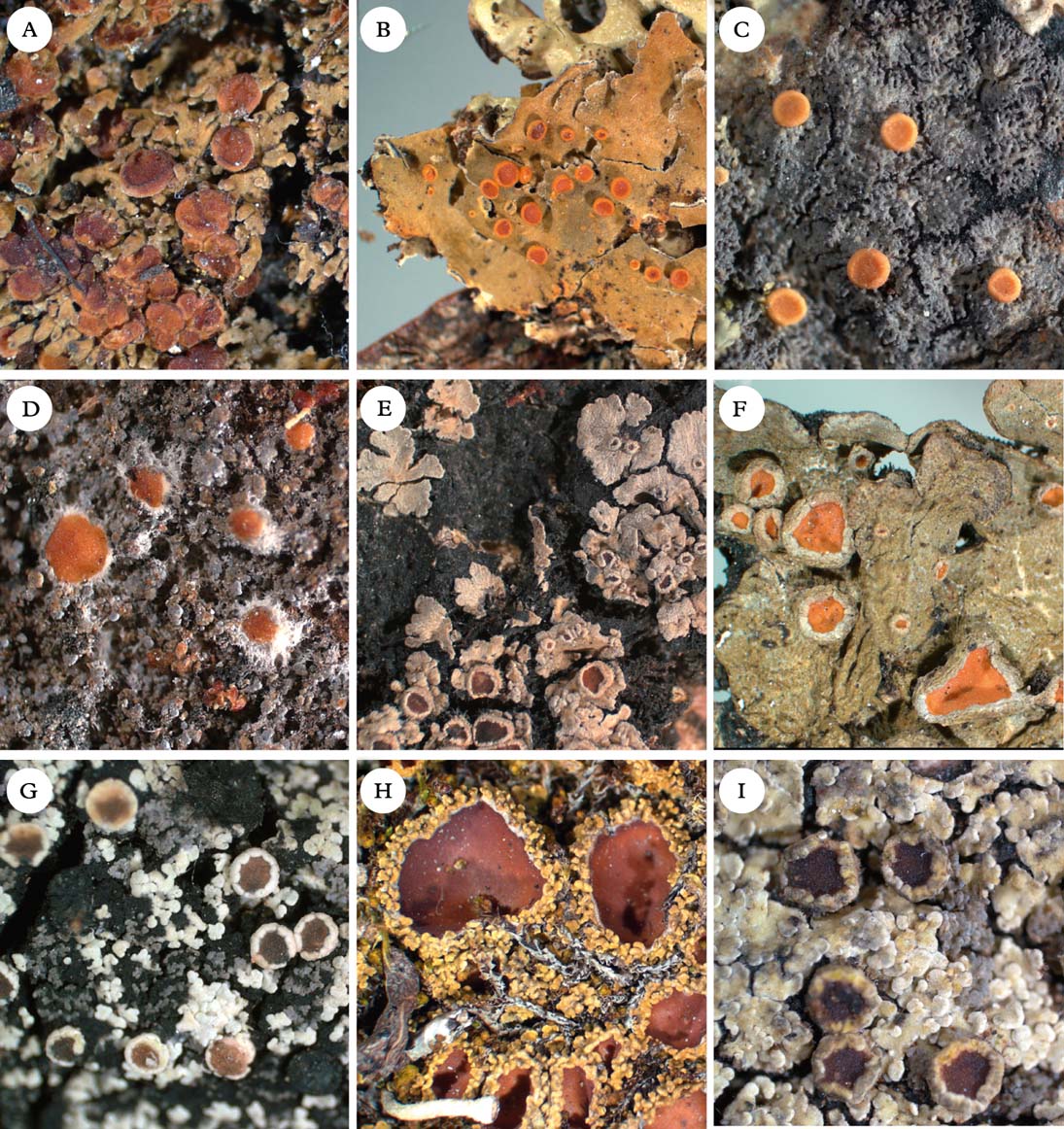
Fig. 5. Representatives of Clade 2c and Xanthopsoroma. A, Nebularia incrassata; B, Fuscoderma applanatum; C, Austroparmeliella lacerata; D, Austrella arachnoidea; E, Lepidocollema marianum; F, Physma byrsaeum; G, Psorophorus pholidotus; H, Psoroma hypnorum; I, Xanthopsoroma contextum. Photographs: Jan Berge. In colour online.
Protopannaria (Gyeln.) P. M. Jørg. & S. Ekman (Fig. 3B) is comprised of seven known crustose-squamulose species without secondary chemistry, apothecia with a thalline margin, and amyloid hymenia with asci lacking internal amyloid structures (Jørgensen Reference Jørgensen2001a , Reference Jørgensen b , Reference Jørgensen2004, Reference Jørgensen2007; Øvstedal & Fryday Reference Øvstedal and Fryday2011). In our phylogeny, P. pezizoides is sister to Santessoniella grisea (Hue) Henssen (Fig. 6D). An undescribed species closely related to Santessoniella crossophylla (Tuck.) P. M. Jørg. (Fig. 6C) is sister to P. pezizoides and S. grisea. Unlike P. pezizoides, the two species of ‘Santessoniella’ have a hemiamyloid hymenium and an internal apical ring structure in the asci. These differences make it unlikely that they can be included in Protopannaria, despite strong branch support in our phylogeny. At the moment, we retain Protopannaria in its current circumscription and refrain from suggesting alternative classifications for the two species of ‘Santessoniella’. We note, however, that relationships and generic boundaries in this group are in need of further study.
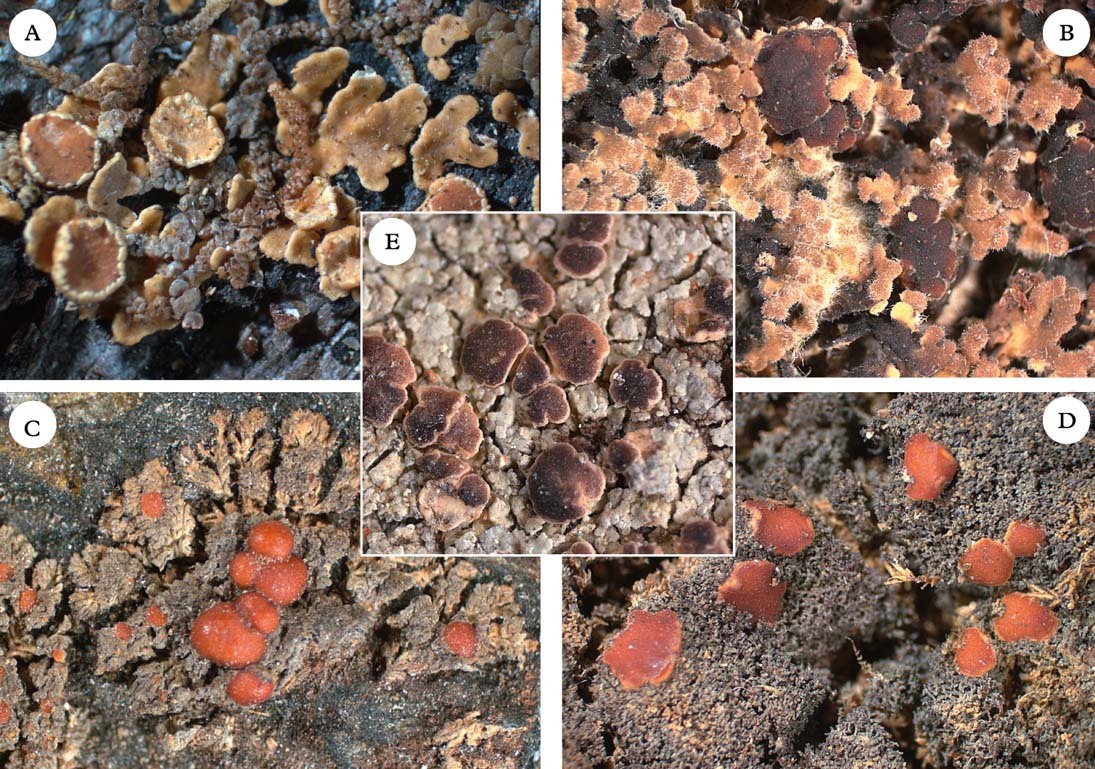
Fig. 6. Five members of the Pannariaceae with uncertain generic affiliation. A, Pannaria isabellina; B, Pannaria hispidula; C, Santessoniella crossophylla; D, Santessoniella grisea; E, Degeliella versicolor. Photographs: Jan Berge. In colour online.
Psoroma Ach. ex Michx (Fig. 5H) traditionally accommodated Pannaria-like species with a green-algal photobiont and a thalline margin surrounding the apothecia. Jørgensen (Reference Jørgensen2001a ) restricted the circumscription of the genus to include close relatives of the type species Psoroma hypnorum (Vahl) Gray, that is, small-squamulose, bryophilous species without lichen substances, and with an amyloid tube- or ring-like structure in the ascus apex. Branch support within Clade 2c in our phylogeny is poor and provides little guidance for revised generic delimitations. We provisionally retain Psoroma more or less as currently understood, with few amendments: ‘Pannaria’ dichroa and ‘P.’ orphnina (along with the two similar species ‘P.’ obscurior and ‘P.’ xanthorioides) are referred here to Psoroma despite their cyanobacterial photobiont, because our phylogeny provides support for their exclusion from Pannaria. Indeed, in accordance with their phylogenetic placement, the asci of these species have a wide amyloid ring structure, which can, however, be difficult to observe. ‘Pannaria’ orphnina is the type species of the genus Siphulina (Hue) C. W. Dodge (Jørgensen Reference Jørgensen2005b ), which accordingly becomes a taxonomic synonym of Psoroma. Furthermore, although Psoroma tenue does not form a monophyletic group with the rest of Psoroma in our phylogeny, there is no support for its exclusion. Chemically, however, P. tenue and its relatives deviate from the rest of Psoroma by producing porphyrilic acid and related substances. We refrain from transferring ‘Santessoniella’ arctophila, sister to P. tenue with high support, to Psoroma or any other genus in the absence of a well-resolved phylogeny. We have, however, chosen to include Santessoniella polychidioides (and its close relative S. macrospora) in Psoroma, because there is reasonable support (0·92 posterior probability) for a close relationship with P. aphthosum and because branch attachment frequencies calculated by Phyutility show that the remaining posterior probability (0·08) is divided between two other positions nested inside our understanding of Psoroma. This choice makes Santessoniella a taxonomic synonym of Psoroma. With these amendments, Psoroma includes species with small-squamulose or rarely small-fruticose thalli with a green-algal or cyanobacterial primary photobiont, and mostly a lack of secondary chemistry (the presence of porphyrilic acid and related substances in Psoroma tenue and relatives being an exception, if included).
Our phylogenetic tree indicates that the widespread P. hypnorum, type species of the genus, is paraphyletic. Further investigations need to determine whether this observation is caused by incomplete lineage sorting or the occurrence of multiple species within P. hypnorum as currently delimited. Psoroma hypnorum specimen III deviates conspicuously from other specimens in having a cyanobacterial (Nostoc) photobiont instead of the standard primary green-algal one (Holien & Jørgensen Reference Holien and Jørgensen2000). The cyanobacterial photobiont confers dramatic modifications to overall lichen morphology towards a growth form similar to taxa currently classified in Santessoniella. Our phylogeny indicates, however, that the fungal component of the cyanobacterial morph is closely related to at least some green-algal representatives (here P. hypnorum specimen V; see Fig. 1). It should also be pointed out that the determination of the P. fruticulosum specimen used to generate the sequences was questioned by Passo et al. (Reference Passo, Stenroos and Calvelo2008).
Psoromaria Nyl. ex Hue may deserve recognition as a genus (see Degeliella). It originally contained two species, P. subdescendens Nyl. (=Degeliella versicolor) and P. descendens Nyl. (=Psoromidium aleuroides). The former was later selected as lectotype (Clements & Shear Reference Clements and Shear1931: 319). Galloway & James (Reference Galloway and James1985) treated both species in Psoromidium Stirt. (as P. aleuroides and P. versicolor), whereas Jørgensen (Reference Jørgensen2004) referred P. versicolor to Degeliella, regarding it as the green counterpart of D. rosulata. In doing so, the older name Psoromaria was unfortunately overlooked. Although we note that Psoromaria may be available for Degeliella versicolor if treated as a separate genus, we refrain from nomenclatural changes at the moment, in anticipation of taxonomical and nomenclatural clarifications in the group.
*Psoromidium Stirt. was reinstated by Galloway (Reference Galloway1983) and Galloway & James (Reference Galloway and James1985) for two species, the type species P. wellingtonii Stirt. [=P. aleuroides (Stirt.) D. J. Galloway] and P. versicolor (Müll. Arg.) D. J. Galloway. The latter was later transferred to the new genus Degeliella (see that genus and Psoromaria). Psoromidium aleuroides is characterized by a thallus of adpressed squamules resting on a distinct hypothallus, a green-algal primary photobiont and distinct cephalodia with Nostoc, an amyloid hymenium, an ascus with an apical ring structure, and a lack of secondary chemistry (Galloway & James Reference Galloway and James1985). Apart from the evanescent apothecial thalline margin in species of Psorophorus (Elvebakk et al. Reference Elvebakk, Robertsen, Park and Hong2010), morphology suggests a close relationship between the two genera. If proven synonymous, Psoromidium is the older name. We provisionally retain Psoromidium, although we note that further studies are needed.
Psorophorus Elvebakk & Hong (Fig. 5G) was recently described by Elvebakk et al. (Reference Elvebakk, Robertsen, Park and Hong2010) for the type species P. pholidotus (Mont.) Elvebakk and P. fuegiensis (Zahlbr.) Elvebakk & Hong. Both species were included in our phylogeny and together form a well-supported monophyletic group sister to Austroparmeliella lacerata. The relationship with Psoromidium needs further study (see that genus).
Ramalodium Nyl. (Fig. 4A) currently comprises six species, R. succulentum Nyl. being the type (Henssen Reference Henssen1965, Reference Henssen1979, Reference Henssen1999). We included only the type species in our phylogeny (as did Wedin et al. Reference Wedin, Wiklund, Jørgensen and Ekman2009). Ramalodium succulentum is recovered as sister to Staurolemma. Ramalodium and Staurolemma have been considered closely related on morphological grounds, the main difference between the genera being the lecideine apothecia in Ramalodium and zeorine apothecia in Staurolemma (Henssen Reference Henssen1999).
Santessoniella Henssen, the type species of which is S. polychidioides (Zahlbr.) Henssen (Fig. 7B), was originally established by Henssen (Reference Henssen1997) for a set of six small, often subfruticose and sometimes gelatinous species with Parmeliella-like apothecia (Henssen Reference Henssen1997). The genus continued to be used in this sense, and another seven species have later been described or transferred to that genus (Jørgensen Reference Jørgensen1998, Reference Jørgensen1999, Reference Jørgensen2005a ; Henssen Reference Henssen2000; Henssen & Kantvilas Reference Henssen and Kantvilas2000; Spribille et al. Reference Spribille, Jørgensen, Schulz and Houde2007; Jørgensen & Palice Reference Jørgensen and Palice2010).

Fig. 7. Type species of two abandoned genera. A, Moelleropsis nebulosa, referred here to Fuscopannaria; B, Santessoniella polychidioides, referred here to Psoroma. Photographs: Jan Berge. In colour online.
Our phylogeny includes five species of Santessoniella, the type species S. polychidioides, S. arctophila, S. saximontana, an undescribed species close to S. crossophylla, and S. grisea. These species are dispersed across much of the tree and constitute the most extreme example of genus-level non-monophyly in our investigation. The type species Santessoniella polychidioides is nested inside Psoroma with moderate support. Morphologically, it may be considered a cyanobacterial expression of a Psoroma, not unlike the cyanobacterial morph of P. hypnorum (Holien & Jørgensen Reference Holien and Jørgensen2000; P. hypnorum III in our tree). It is noteworthy, however, that the asci of S. polychidioides and relatives are more narrowly cylindrical than in Psoroma s. str., with a tube-like amyloid internal structure as opposed to the wider ring-like structure in Psoroma s. str. In addition, the hymenial reaction is more pronouncedly hemiamyloid in S. polychidioides and relatives, rapidly changing from blue-green to red-brown, whereas in Psoroma s. str. the reaction is blackish blue, turning slowly to sordid blue. Santessoniella saximontana is nested inside Leciophysma with high support and seems to share morphological characteristics of that genus (Henssen Reference Henssen1965). Santessoniella grisea and the undescribed relative of S. crossophylla are closely related to Protopannaria, from which they differ markedly with respect to morphology. Finally, S. arctophila seems to be closely related to Psoroma tenue. Their relationships remain unclear and we refrain here from assigning them to a genus.
Siphulastrum Müll. Arg. (Fig. 2F) is a genus of four species, one of which is the type species S. triste Müll. Arg. (Jørgensen Reference Jørgensen2003). The genus is characterized by a heteromerous thallus with a Scytonema photobiont, a hemiamyloid hymenial reaction, lack of apical structures in the asci, presence of argopsin in the thallus, and a dense upper cortex of incrassate cells with small cell lumina. Unfortunately, material of the type species itself was not available for our study, although the included species, S. squamosum, conforms to the generic characteristics and is likely to be closely related to the type species. In our phylogenetic tree, Siphulastrum is the sister group to Leioderma and Degeliella rosulata.
Staurolemma Körb. (Fig. 4B) includes eight known species (Jørgensen Reference Jørgensen2010) and is typified by S. dalmaticum Körb., a synonym of S. omphalarioides (Anzi) P. M. Jørg. & Henssen. We included two species in our phylogeny, which form a monophyletic group with high support. Furthermore, Staurolemma is the sister group to Ramalodium in our phylogeny as well as that of Wedin et al. (Reference Wedin, Wiklund, Jørgensen and Ekman2009). This corroborates the view that the two genera are closely related on morphological grounds, differing mainly in apothecial anatomy (Henssen Reference Henssen1999).
Note that ‘Staurolemma sp. nov.’ included in the phylogeny of Wedin et al. (Reference Wedin, Wiklund, Jørgensen and Ekman2009) has been described as S. oculatum P. M. Jørg. & Aptroot (Jørgensen Reference Jørgensen2010).
*Steineropsis T. Sprib. & Muggia was described for the single species S. alaskana T. Sprib. & Muggia by Spribille et al. (Reference Spribille, Pérez-Ortega, Tønsberg and Schirokauer2010). This species superficially resembles a Placopsis and the thallus is characterized by a paraplectenchymatous upper cortex, which extends into the medulla. Apothecia and pycnidia have not been described. Steineropsis alaskana was sister to Protopannaria in the phylogeny of Spribille & Muggia (Reference Spribille and Muggia2013).
Xanthopsoroma Elvebakk & Hong (Fig. 5I) was established to accommodate the type species X. contextum (Stirt.) Elvebakk and X. soccatum (R. Br. ex Crombie) Elvebakk, two Southern Hemispheric species previously treated in Psoroma and containing usnic acid and a series of terpenoids (Elvebakk et al. Reference Elvebakk, Robertsen, Park and Hong2010). Support for its monophyly in our phylogeny is poor. Surrounding branches have high support, but we cannot exclude the possibility that Xanthopsoroma is paraphyletic. However, at least one, possibly both members of the genus are likely to be sister to Clade 2a–c (Fig. 1).
Provisional key to the genera
-
1 Thallus gelatinous, mostly without lichen substances (PD−) ... 2
Thallus not gelatinous, often with lichen substances (PD+) ... 11
-
2(1) Thallus subfruticose to fruticulose, sometimes nearly granular ... 3
Thallus squamulose to foliose ... 5
-
3(2) Thallus applanate, finely and dichotomously dissected; photobiont Scytonema; medullary hyphae parallel to cortex; tropical ... Leptogidium
Thallus erect, consisting of coarser and often irregular branches; photobiont Nostoc; medullary hyphae at an angle to the cortex, usually in a reticulate pattern temperate ... 4
-
4(3) Lobes up to 0·3 mm wide, sometimes nearly granular; hyphal walls distinctly gelatinized ... Leciophysma
Lobes up to 1 mm wide, more or less squamulose; hyphal walls not or weakly gelatinized ... Psoroma pro parte (‘Santessoniella’ s. str.)
-
5(2) Apothecia without thalline margin; thallus mostly squamulose or nearly subfruticose; Southern Hemisphere ... 6
Apothecia with thalline margin; thallus with wider, flattened lobes, subfoliose to foliose; tropical ... 7
-
6(5) Thallus membranaceous; excipulum annular; asci with internal apical amyloid ring or tube ... Homothecium
Thallus squamulose (to subfruticose); excipulum cupular; asci without internal apical amyloid structures ... Ramalodium
-
7(5) Thallus with fan-shaped lobes, tawny, with pannarin (PD+); asci without internal apical amyloid structures ... Pannaria lurida group
Thallus with narrow, elongated lobes, bluish grey, without pannarin (PD−); asci with internal apical amyloid ring structures ... 8
-
8(7) Thallus homoiomerous, containing terpenoids; ascospores globose, faintly brownish ... Kroswia
Thallus heteromerous, without secondary substances; ascospores ellipsoid, colourless ... 9
-
9(8) Thallus resting on a distinct mat of protruding blackish rhizohyphae; cortex cellular, one-layered; Brazil ... Lepidocollema carassense
Thallus without protruding rhizohyphae; cortex multi-layered; paleotropical ... 10
-
10(9) Thallus with narrow, elongated lobes; cortex of 1–3 cell layers; apothecia stipitate without supportive tissue; Sri Lanka ... Leightoniella zeylanica
Thallus with wider lobes; cortex of densely agglutinated hyphae; apothecia sessile with supportive tissue; widespread in the tropics ... Physma
-
11(1) Thallus with green-algal photobiont ... 12
Thallus with cyanobacterial photobiont ... 19
-
12(11) Asci without internal amyloid apical structures ... 13
Asci with internal amyloid apical structures ... 14
-
13(12) Thallus squamulose-foliose, with pannarin and related substances (PD+); apothecia with thalline margin ... Pannaria
Thallus of closely adpressed squamules, without secondary substances; apothecia biatorine, without thalline margin ... Degeliella versicolor
-
14(12) Squamules with a yellow tinge, with usnic acid ... Xanthopsoroma
Squamules without a yellow tinge, without usnic acid ... 15
-
15(14) Apothecia often proliferating, without thalline margin; thallus with cottony prothallus ... Psoromidium
Apothecia single, with distinct thalline margin; thallus without cottony prothallus ... 16
-
16(15) Thallus with pannarin (PD+); asci with distinct apical amyloid cap-shaped plug ... Joergensenia cephalodina
Thallus without pannarin (PD−); asci with amyloid apical ring structures ... 17
-
17(16) Apothecia flat, often with convex, dark brown disc ... Fuscopannaria viridescens
Apothecia urceolate with concave, light or orange-brown disc ... 18
-
18(17) Squamules appressed, resting on a distinct blackish prothallus; corticolous ... Psorophorus
Squamules loosely scattered over the substratum, without prothallus; usually bryophilous or terricolous ... Psoroma
-
19(11) Thallus of closely appressed, chestnut brown squamules with cream-coloured soralia; hypothallus blue-black; photobiont Nostoc; nearly always sterile; Atlantic-Mediterranean ... Nevesia sampaiana
Thallus with different combination of characters ... 20
-
20(19) Thallus a small, placodioid, Placopsis-like rosette with opuntioid lobules and a thick, paraplectenchymatous upper cortex; sterile ... Steineropsis alaskana
Thallus with different combination of characters ... 21
-
21(20) Apothecia with secondary thalline margin ... 22
Apothecia without secondary thalline margin, or rarely with thalline corona ... 25
-
22(21) Thallus on distinct blackish hypothallus, without lichen substances or pigments; tropical ... Lepidocollema
Thallus not on distinct hypothallus, often with lichen substances or pigments ... 23
-
23(22) Thallus squamulose-foliose, usually with pannarin (PD+); asci without internal, apical amyloid structures ... Pannaria
Thallus small-squamulose, usually without pannarin (PD−), rarely with pannarin (but then also with argopsin); asci with internal amyloid ring structures ... 24
-
24(23) Apothecia with distinct thalline margin, disc orange-brown, hymenium amyloid ... Protopannaria
Apothecia with variably developed thalline margin, disc brown to blackish; hymenium hemiamyloid ... Fuscopannaria
-
25(21) Thallus foliose, not closely appressed to substratum, upper surface usually with hairs ... 26
Thallus squamulose or placodioid, often closely appressed to substratum, upper surface without hairs ... 27
-
26(25) Thallus always hairy, often with stiff prominent hairs; apothecia usually marginal and stalked, if otherwise always PD+ orange (eriodermin) ... Erioderma
Thallus arachnoid-tomentose or glabrous, with laminal, sessile apothecia, PD− ... Leioderma
-
27(25) Thallus placodioid, often forming large circular blue-grey thalli with well-developed bluish black rhizohyphae ... 28
Thallus not placodioid, without or with brownish rhizohyphae ... 31
-
28(27) Hymenium non-amyloid; asci without internal apical amyloid structures; photobiont Scytonema ... Degeliella rosulata
Hymenium amyloid; asci with internal apical amyloid structures; photobiont Scytonema or Nostoc ... 29
-
29(28) Thallus thin and Coccocarpia-like, with a Scytonema photobiont; upper cortex prosoplectenchymatous, consisting of a few cell layers of periclinally arranged hyphae; Southern Hemisphere ... Degelia sect. Degelia
Thallus thick and rigid; upper cortex paraplectenchymatous, consisting of several layers of anticlinally arranged hyphae ... 30
-
30(29) Photobiont Nostoc; thallus losely attached to substratum, with upper surface having prominent longitudinal ridges; Northern Hemisphere ... Pectenia
Photobiont Scytonema, thallus appressed to substratum, with smooth upper surface; Subantarctic ... Degelia sect. Frigidae
-
31(27) Apothecia surrounded by a thin weft of hyphae; asci narrowly elongate, thin-walled, without internal apical structures ... Austrella
Apothecia not surrounded by a weft of hyphae; asci wider, clavate to cylindrical, thick-walled, with internal apical amyloid structures ... 32
-
32(31) Apothecia with thick proper exciple, containing elements of the formative supporting tissue and photobiont cells penetrating into the subhymenium ... 33
Apothecia with thin proper exciple, without supporting tissue or photobiont cells in the subhymenium ... 34
-
33(32) Lower surface with small, curled brownish hairs; hymenium hemiamyloid; Southern Hemisphere ... Fuscoderma
Lower surface without brownish hairs; hymenium amyloid; Andean Mountains ... Nebularia
-
34(32) Thallus thick and often shiny, containing argopsin (PD+); cortical cells thick-walled; hymenium hemiamyloid; mostly terricolous ... Siphulastrum
Thallus thin and usually dull, without argopsin (PD−); cortical cells thin-walled; hymenium amyloid; mostly corticolous ... 35
-
35(34) Thallus often forming both upper and lower cortex; subhymenium flat, colourless ... Austroparmeliella
Thallus only with upper cortex; subhymenium often lentil-shaped, brownish ... Parmeliella
New Species-level Combinations
In this section, we list species-level combinations necessitated by the generic classification proposed above.
Austroparmeliella chilensis (Hue) P. M. Jørg. comb. nov.
MycoBank No.: MB807985
Placynthium chilense Hue, Bull. Soc. Linn. Normandie ser. 5, 9: 158–159 (1906); type: Chile, Cordillera de Ranco, Lechler 3016 (PC—lectotype! fide Henssen Reference Henssen1997: 83).
Austroparmeliella elongata (Henssen) P. M. Jørg. comb. nov.
MycoBank No.: MB807986
Santessoniella elongata Henssen, Symb. Bot. Ups. 32: 83–84 (Reference Henssen1997); type: Argentina, Rio Negro, Parque Nacional Nahuel Huapi, Puerto Blest, 1973, Vobis & Henssen 24606a (H—holotype!).
Austroparmeliella lacerata (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB807987
Parmeliella lacerata P. M. Jørg., Lichenologist 30: 537 (1998); type: Republic of South Africa, Western Cape, Riviersonderend, Oubos Forest, 1996, Nordin 4542 (UPS—holotype!).
Austroparmeliella rakiurae (P. M. Jørg. & D. J. Galloway) P. M. Jørg. comb. nov.
MycoBank No.: MB807988
Parmeliella rakiurae P. M. Jørg. & D. J. Galloway, Bibl. Lich. 88: 244 (2004); type: New Zealand, Stewart Island, Port Pegasus, track from Disappointment Cove to Broad Bay, coastal forest on Olearia bark, 29 July 2001, Galloway 0845 (BG—holotype!).
Austroparmeliella rosettiformis (Henssen) P. M. Jørg. comb. nov.
MycoBank No.: MB807989
Santessoniella rosettiformis Henssen, Lichenologist 32: 18 (Reference Henssen and Kantvilas2000); type: Chile, Prov. Arauco, Parque Nacional Contulma, 1973, Vobis & Henssen 24281 (H—holotype!).
Leciophysma saximontana (T. Sprib. et al.) P. M. Jørg., Wedin & S. Ekman comb. nov.
MycoBank No.: MB807990
Santessoniella saximontana T. Sprib., P. M. Jørg. & M. Schulz, Bibl. Lich. 98: 288 (2007); type: Canada, British Columbia, Rocky Mts., Albert River drainage, 2006, Spribille 21173 & Houde (CANL—holotype!).
Lepidocollema adpressum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB807991
Parmeliella adpressa P. M. Jørg., Journ. Japanese Bot. 76: 289 (2001); type: Japan, Shinano Prov., Mt. Kinpu, alt. 2100–2450 m, Jinzenji 176 (TNS—holotype!).
Lepidocollema allochroum (Makhija & Adaw.) P. M. Jørg. comb. nov.
MycoBank No.: MB807992
Parmeliella allochroa Makhija & Adawad, Mycotaxon 71: 327 (1999); type: India, Nicobar Islands, Car Nicobar, Kimus, on coconut palm, 29 December 1986, Patwardhan & Sethy 86.787 (AMH—holotype!).
Lepidocollema borbonicum (P. M. Jørg. & Schumm) P. M. Jørg. comb. nov.
MycoBank No.: MB807993
Parmeliella borbonica P. M. Jørg. & Schumm, Lichenologist 42: 697 (2010); type: La Réunion, Takamaka, low mountain forest near the electrostation, alt. 790 m, 10 September 2009, Frahm & Schumm (BG—holotype!).
Lepidocollema brisbanense (C. Knight) P. M. Jørg. comb. nov.
MycoBank No.: MB807994
Pannaria brisbanensis C. Knight in J. Shirley, Proc. Royal Soc. Queensland 6: 194 (1890); type: Australia, Queensland, Brisbane River, Shirley 113 (WELT—lectotype! fide Jørgensen & Galloway Reference Jørgensen and Galloway1992).
Lepidocollema cineratum (Zahlbr.) P. M. Jørg. comb. nov.
MycoBank No.: MB807995
Pannaria cinerata Zahlbr. in H. Magn. & Zahlbr., Ark. Bot. 31A(1): 72 (1943); type: USA, Hawaii, Kauai, Kauhao, 1000 m, 8 February 1910, Faurie 274 (W—holotype!).
Lepidocollema endoluteum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB807996
Parmeliella endolutea P. M. Jørg., Lichenologist 39: 239 (2007); type: Philippines, Prov. Rizal, Mt. Irid, Herre (LAM—holotype!).
Lepidocollema endomiltum (Vain.) P. M. Jørg. comb. nov.
MycoBank No.: MB807997
Pannaria endomilta Vain., Ann. Acad. Sci. Fenn. ser. A 15, 6: 15 (1921); type: Philippines, Mindanao, Davao, Mt. Apo, 6000 ft., 21 April 1904, Copeland 1090 p.p. (TUR-V 12168—holotype!).
Lepiodocollema exornatum (Zahlbr.) P. M. Jørg. comb. nov.
MycoBank No.: MB807998
Pannaria cinerata var. exornata Zahlbr. in H. Magn. & Zahlbr., Ark. Bot. 31A(1): 72 (1943); type: USA, Hawaii, Maui, Hana, September 1909, Faurie 559 (W—lectotype! fide Jørgensen Reference Jørgensen2003).
Lepidocollema fuscatum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB807999
Parmeliella fuscata P. M. Jørg., Bryologist 106: 123 (2003); type: India, Maharasta, Ambenali, on road to Pratapgad, felled tree at bridge, 2 November 1973, Hale 40045 (US—holotype!).
Lepidocollema granuliferum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808000
Parmeliella granulifera P. M. Jørg., Bibl. Lich. 78: 129 (2001); type: Australia, Northern Territory, Channel Island, prawn farm, landward edge of mangroves, 1991, Benfield (BRI—holotype!).
Lepidocollema imbricatulum (Müll. Arg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808001
Pannaria imbricatula Müll. Arg., Flora 64: 507 (1881); type: Brazil, São Paolo, prope Apiahy, Puiggari 148 p.p. (G—holotype!).
Lepidocollema leiostroma (Nyl.) P. M. Jørg. comb. nov.
MycoBank No.: MB808002
Pannaria leiostroma Nyl. in Leight., Trans. Linn. Soc. London 27: 165 (1869); type: Sri Lanka, Thwaites (BM—holotype!).
Lepidocollema macrosporum (Makhija & Adawad.) P. M. Jørg. comb. nov.
MycoBank No.: MB808003
Parmeliella macrospora Makhija & Adawad., Mycotaxon 71: 332 (1999); type: India, Nicobar Island, Car Nicobar, Kimus, 1986, Sethy & Nagarkar 86.778 (AMH—holotype!).
Lepidocollema marianum (Fr.) P. M. Jørg. comb. nov.
MycoBank No.: MB808004
Parmelia mariana Fr., Syst. Orb. Veg. I: 284 (1825); type: Mariana Islands, Gaudichaud (UPS—holotype!).
Lepidocollema montanum (P. M. Jørg. & Sipman) P. M. Jørg. comb. nov.
MycoBank No.: MB808005
Parmeliella montana P. M. Jørg. & Sipman, J. Hattori Bot. Lab. 100: 711 (2006); type: Papua New Guinea, Morobe Distr., Mt. Kaindi, Wau, 1973, Kashiwadani 10541 (TNS—holotype!).
Lepidocollema nitidum (P. M. Jørg. & Sipman) P. M. Jørg. comb. nov.
MycoBank No.: MB808006
Parmeliella nitida P. M. Jørg. & Sipman, J. Hattori Bot. Lab. 100: 712 (2006); type: Papua New Guinea, Simbu Prov., Mt. Wilhelm, Pindaunde valley, near hut on S-shore of Lake Piunde, W slope of valley, c. 3700 m, 6 August 1992, Sipman 35688 (B—holotype!).
Lepidocollema pannarioides (P. M. Jørg. & Sipman) P. M. Jørg. comb. nov.
MycoBank No.: MB808007
Parmeliella pannaroides P. M. Jørg. & Sipman, J. Hattori Bot. Lab. 100: 713 (2006); type: Papua New Guinea, Western Highland Distr., Mt. Wilhelm, en route from Kombugomanubo to the Pindaunde Lakes, 1974, Kashiwadani 11064 (TNS—holotype!).
Lepidocollema papillatum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808008
Parmeliella papillata P. M. Jørg., Bibl. Lich. 78: 133 (2001); type: Australia, Queensland, Cardwell Range, 45 km NW of Cardwell, 1986, Elix 20169 & Streimann (CANB—holotype!).
Lepidocollema polyphyllinum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808009
Parmeliella polyphyllina P. M. Jørg., Bibl. Lich. 78: 134 (2001); type: Australia, Queensland, Hugh Nelson Range, along Plath Road, 15 km S of Atherton, alt. 1080 m, 25 June 1984, Elix 16363 & Streimann (CANB—holotype!).
Lepidocollema stylophorum (Vain.) P. M. Jørg. comb. nov.
MycoBank No.: MB808010
Pannaria stylophora Vain., Add. Lichenogr. Antill.: 102 (1915); type: Guadeloupe, Sofaga, ad corticem Sapii aucupari, Duss 1387 (TUR-V 12107—holotype!).
Lepidocollema wainioi (Zahlbr.) P. M. Jørg. comb. nov.
MycoBank No.: MB808011
Pannaria wainioi Zahlbr., Cat. Lich. Univ. 3: 261 (1924); type: Philippines, Mindanao, Butuan, alt. 15 m, 1911, Weber 1387 (TUR-V 12178—lectotype! fide Jørgensen Reference Jørgensen2003).
Lepidocollema zeylanicum (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808012
Parmeliella zeylanica P. M. Jørg., Lichenologist 41: 257 (2009); type: Sri Lanka, Nuwara Elyia, near the Golf Club, 1964, Degelius As-438 (UPS—holotype!).
Nebularia incrassata (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808013
Parmeliella incrassata P. M. Jørg., Lichenologist 32: 141 (2000); type: Ecuador, Pichincha, eastern slopes of Cerro Iliniza, alt. 4200 m, epiphyte on Polylepis, 7 March 1972, Arvidsson & Nilsson (GB—holotype!).
Nebularia psoromoides (P. M. Jørg. & Palice) P. M. Jørg. comb. nov.
MycoBank No.: MB808014
Parmeliella psoromoides P. M. Jørg. & Palice, Nordic J. Bot. 28: 625 (2010); type: Ecuador, Carchi, Volcan Chiles, páramo c. 1·5 km south-west of the top, 4050 m a.s.l., 13 July 1999, Palice 11977 (PRA—holotype!).
Nevesia sampaiana (Tav.) P. M. Jørg., L. Lindblom, Wedin & S. Ekman comb. nov.
MycoBank No.: MB808015
Pannaria sampaiana Tav., Port. Acta Biol. ser. B 3: 76–77 (1950); type: Portugal, Minho, Serra do Gerês, between S. Bento da Porta Alberta and Freitas, 1948, Tavares 2829 (LISU—holotype!).
Pectenia atlantica (Degel.) P. M. Jørg., L. Lindblom, Wedin & S. Ekman comb. nov.
MycoBank No.: MB808016
Parmeliella atlantica Degel., Acta Phytogeogr. Suec. 7: 131 (1935); type: Ireland, Killarney, near Muckross lake, 1933, Degelius (UPS—holotype!).
Pectenia cyanoloma (Schaer.) P. M. Jørg., L. Lindblom, Wedin & S. Ekman comb. nov.
MycoBank No.: MB808017
Parmelia plumbea var cyanoloma Schaer., Enum. Criticae Lich. Eur.: 36 (1850); type: France, Normandie, in sylva Briquebec, Delise (G—lectotype! fide Jørgensen Reference Jørgensen1978).
Pectenia ligulata (P. M. Jørg. & P. James) P. M. Jørg., L. Lindblom, Wedin & S. Ekman comb. nov.
MycoBank No.: MB808018
Degelia ligulata P. M. Jørg. & P. James, Bibl. Lich. 38: 266 (1990); type: Portugal, Azores, Faial, Santa Maria, Anjos, on mossy earth bank, 1976, James (BM—holotype!).
Pectenia plumbea (Lightf.) P. M. Jørg., L. Lindblom, Wedin & S. Ekman comb. nov.
MycoBank No.: MB808019
Lichen plumbeus Lightf., Fl. Scot. 2: 826 (1777); type: Great Britain (OXF-DILL 179: 73a—lectotype! fide Jørgensen Reference Jørgensen1978).
Psoroma dichroum (Hook. f. & Taylor) P. M. Jørg. comb. nov.
MycoBank No.: MB808020
Lecanora dichroa Hook.f. & Taylor, J. Bot. London 3: 643 (1844); type: Kerguelen, Christmas harbour, Hooker 1844 (FH—lectotype! fide Dodge Reference Dodge1948).
Psoroma macrosporum (P. M. Jørg. & Palice) P. M. Jørg. comb. nov.
MycoBank No.: MB808021
Santessoniella macrospora P. M. Jørg. & Palice, Nord. J. Bot. 28: 626 (2010); type: Ecuador, Carchi: surroundings of Laguna Verde, 1·5–1·8 km SSE of Volcan Chiles, 4000 m a.s.l., 12 July 1999, Palice 2750 (PRA—holotype!).
Psoroma obscurius (Nyl.) P. M. Jørg. comb. nov.
MycoBank No.: MB808022
Pannaria obscurior Nyl. in Cromb., J. Bot. London 13: 334 (1875); type: Kerguelen, Observatory Bay, December 1874, Eaton (BM—holotype!).
Psoroma orphninum (Hue) P. M. Jørg. comb. nov.
MycoBank No.: MB808023
Siphula orphnina Hue, 2me Exped. Antarct. FranV., Lich.: 19 (1915); type: South Shetland, Livingston Island, South Bay, Johnson's Peak, 300–350 m, 8 February 1998, Søchting 7833 (BG—neotype! fide Jørgensen Reference Jørgensen2005b ).
Psoroma polychidioides (Zahlbr.) P. M. Jørg. comb. nov.
MycoBank No.: MB808024
Lemmopsis polychidioides Zahlbr. in Skottsb.: Nat. Hist. of Juan Fernandez Easter Isl. 2: 333 (1924); type: Chile, Juan Fernandez, Mastierra, Cordon Chifladores, 17 April 1917, Skottsberg & Skottsberg 408 (UPS—holotype!).
Psoroma xanthorioides (P. M. Jørg.) P. M. Jørg. comb. nov.
MycoBank No.: MB808025
Pannaria xanthorioides P. M. Jørg., Bibl. Lich. 88: 238 (2004); type: Heard Island, near the summit of Scarlet Hill, 6 January 2001, Gremmen H-0663 (CANB—holotype!).
We gratefully acknowledge the following herbaria for assistance: AMH, B, BG, BM, BRI, CANB, CANL, FH, G, GB, H, LAM, LISU, OXF, PC, PRA, TNS, TUR, UPS, US, W, and WELT. We are indebted to Frances Anderson, André Aptroot, Franz Berger, Pieter van den Boom, William R. Buck, Arve Elvebakk, Javier Etayo, David J. Galloway, Dag Olav Øvstedal, Zdenek Palice, Mac Pitcher, Felix Schumm, Harrie Sipman, Ulrik Søchting, Tor Tønsberg, Dalip K. Upreti, as well as the curators of BG, NBM, NMW, PRA, S, and UPS for providing us with material that was used to obtain sequence data. We thank Birgit Kanz, Heidi Lie Andersen, and the staff at the Molecular Systematics Laboratory at the Swedish Museum of Natural History for assisting with laboratory work. Special thanks are due to Astri Botnen for help in handling herbarium material, Jan Berge for taking the colour photographs, and Beate Helle for arranging the photographs into complete illustrations. This work was supported by the Research Council of Norway (grant no. 128388/420 to SE), the Swedish Research Council (grants VR 621-2006-3760, VR 621-2009-5372, and VR 621-2012-3990 to MW), and the University Museum of Bergen (Olaf Grolle Olsens legat to PMJ).
Supplementary Material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S002428291400019X









