Introduction
There is no consensus on how to best measure the success of species reintroductions (Seddon, Reference Seddon1999; Estrada, Reference Estrada2014), although they are generally considered successful if they result in a population that is viable and self-sustaining (Griffith et al., Reference Griffith, Scott, Carpenter and Reed1989; Caughley & Gunn, Reference Caughley and Gunn1996; Dodd, Reference Dodd and Lannoo2005) or reaches carrying capacity in the release area (Sarrazin, Reference Sarrazin2007). The time required for the establishment of a self-sustaining population varies across species (Seddon, Reference Seddon1999; Fischer & Lindenmayer, Reference Fischer and Lindenmayer2000; Albrecht et al., Reference Albrecht, Guerrant, Maschinski and Kennedy2011), and several demographic parameters and stochastic processes need to be assessed to determine whether a population is likely to be viable in the longer term (Sarrazin & Barbault, Reference Sarrazin and Barbault1996). In the case of long-lived species such as medium-sized and large mammals, this time is often longer than stakeholders allow for, or than funding is available for monitoring (Sutton, Reference Sutton2015). Therefore, although essential for assessing the efficacy of reintroduction projects of most vertebrates (Semlitsch, Reference Semlitsch2002; Muths & Dreitz, Reference Muths and Dreitz2008; IUCN/SSC, 2013), long-term monitoring is rarely achieved (Sutherland, Reference Sutherland2000; Estrada, Reference Estrada2014; but see Mason & Macdonald, Reference Mason and Macdonald2004 and Koelewijn et al., Reference Koelewijn, Pérez-Haro, Jansman, Boerwinkel, Bovenschen and Lammertsma2010 regarding the Eurasian otter Lutra lutra).
The release phase of reintroductions is affected by financial and logistical constraints, and managers' decisions (Converse et al., Reference Converse, Moore and Armstrong2013). In recent decades, the assessment of the size of the founder stock and duration of the release period have been increasingly facilitated by population modelling (Armstrong & Seddon, Reference Armstrong and Seddon2008; King et al., Reference King, Chamberlan and Courage2013), but the absence of decision analysis (i.e. analysis of management objectives and alternative actions to reach them) may have limited the success of previous reintroduction projects (Converse et al., Reference Converse, Moore and Armstrong2013).
The Eurasian otter is a semi-aquatic mustelid whose populations suffered a sharp decline throughout Europe in the 20th century (Macdonald & Mason, Reference Macdonald and Mason1994; Ruiz-Olmo et al., Reference Ruiz-Olmo, Lafontaine, Prigioni, López-Martín and Santos-Reis2000). In the second half of the 20th century, the otter became a symbol for nature conservation. In an attempt to prevent its seemingly unavoidable extinction in several countries, the species was included in the European breeding programme for self-sustaining captive populations (Europäisches Erhaltungszuchtprogramm, EEP), involving zoological gardens and specialized breeding centres (Vogt, Reference Vogt1995). Since the 1980s, the otter has been translocated (UK: Wayre, Reference Wayre1985; Roche et al., Reference Roche, Harris, Warrington and Copp1995; Spain: Fernández-Morán et al., Reference Fernández-Morán, Saavedra and Manteca-Vilanova2002) or reintroduced in several countries (Sweden: Sjöåsen, Reference Sjöåsen1996; The Netherlands: Koelewijn et al., Reference Koelewijn, Pérez-Haro, Jansman, Boerwinkel, Bovenschen and Lammertsma2010; France: Geboes et al., Reference Geboes, Rosoux, Lemarchand, Hansen and Libois2016), and further projects are underway (Denmark: Christiansen, Reference Christiansen2017; Japan: Murakami, Reference Murakami2017).
At the end of the 20th century, the Eurasian otter became extinct in the northern and central Italian peninsula (Prigioni et al., Reference Prigioni, Balestrieri and Remonti2007). On the River Ticino (northern Italy, Piedmont/Lombardy regions), one of the best conserved lowland riverine habitats of northern Italy, the species was declared extinct in 1985 (Cassola, Reference Cassola1986; Prigioni, Reference Prigioni and Cassola1986), after the last reliable signs of its presence had been recorded in 1980 (Galeotti, Reference Galeotti1981). The river was later identified as a potential reintroduction area in the Action Plan for the Conservation of European Otters (Macdonald & Mason, Reference Macdonald, Mason, Foster-Turley, Macdonald and Mason1990), and deemed suitable for otters by a feasibility study (Prigioni, Reference Prigioni1995). In the EEP framework, two breeding centres were built and a reintroduction trialled with the release of one pair of otters (Montanari & Boffino, Reference Montanari and Boffino2000).
Concern about the reintroduction was raised by the Italian Institute for Wildlife, because analyses of the mitochondrial DNA of the otters hosted in European breeding centres confirmed that individuals of the Asiatic subspecies Lutra lutra barang had contributed to the gene pool of the EEP population (the so-called genetic B-line; Randi et al., Reference Randi, Davoli and Pierpaoli2001). In addition, although the European River Otter Studbook was established with the aim to increase the genetic variability of captive-bred otters, a high level of inbreeding was recorded (Randi et al., Reference Randi, Davoli and Pierpaoli2001, Reference Randi, Mucci, Arrel, Bailey, Bonder and Dallas2005).
The reintroduction was interrupted soon after the first release in 1997, mainly because of concerns about the genetic make-up of the captive population. Nonetheless, a second pair of animals was released or escaped in 1998 and a further female with her cub probably escaped before 2000. None of these otters were equipped with radio transmitters; their fate is thus unknown and the precise number of reintroduced animals is uncertain.
Although translocations of large numbers of individuals can have negligible effects on local populations (Arrendal et al., Reference Arrendal, Walker, Sundqvist, Hellborg and Vilà2004), and some reintroductions have been successful with relatively few founders (lynx Lynx lynx: Breitenmoser et al., Reference Breitenmoser, Breitenmoser-Würsten and Capt1998; fisher Martes pennanti: Aubry & Lewis, Reference Aubry and Lewis2003; Eurasian badger Meles meles: Balestrieri et al., Reference Balestrieri, Tirozzi, Agostani and Saino2018), success can be affected by the number of released animals (Griffith et al., Reference Griffith, Scott, Carpenter and Reed1989). Small founder populations are vulnerable to both demographic and environmental stochasticity (Gilpin & Soulé, Reference Gilpin, Soulé and Soulé1986) and thus at risk of local extinction. Nevertheless, since 2007 signs of otter presence such as roadkill and spraints have been recorded regularly around a short stretch (5–7 km) of the Ticino River near the release site (Balestrieri et al., Reference Balestrieri, Remonti, Prigioni and Angelici2016). In 2016–2017 otters were recorded by camera traps near the release site on three occasions, and otter signs were found in 16 out of 32 sampling locations (600 m long river stretches) surveyed (Smiroldo et al., Reference Smiroldo, Balestrieri, Pini and Tremolada2019). Throughout 2018, a total of 77 km of watercourses were monitored, but only five otter spraints were found, in four surveyed stretches of the river (Tremolada et al., Reference Tremolada, Smiroldo, Verduci, Gatti, Boggioni and Gianfranceschi2020). Compared to the core area of the Italian otter range, marking density around the release site has generally been low (0.15–0.2 vs 3.7 spraints/100 m), suggesting that the local otter population probably consists of few individuals (Prigioni et al., Reference Prigioni, Balestrieri, Remonti, Sgrosso and Priore2006a).
The persistence of the species in the release area despite the small founder population also raised questions about the possible survival of a residual native otter population that was not recorded during the last national census (Prigioni, Reference Prigioni and Cassola1986). To assess the abundance of the otter population on the River Ticino and examine the genetic origin of the reintroduced population, we analysed DNA from faecal samples. This method has proved to be effective for assessing the minimum number of individuals and haplotypes of elusive species such as the Eurasian otter (Dallas et al., Reference Dallas, Coxon, Sykes, Chanin, Marshall and Carss2003; Hung et al., Reference Hung, Li and Lee2004; Kalz et al., Reference Kalz, Jewgenow and Fickel2006; Koelewijn et al., Reference Koelewijn, Pérez-Haro, Jansman, Boerwinkel, Bovenschen and Lammertsma2010). In addition, we used population viability analysis to determine a posteriori the number of founders and predict the likely fate of the population.
Methods
Genetic analyses
During the 2016–2018 monitoring surveys, which covered the entire Italian stretch of the River Ticino, we collected 25 sufficiently fresh otter spraints and preserved them in ethanol at −20 °C for genetic analyses (Bonesi et al., Reference Bonesi, Hale and Macdonald2013; Vergara et al., Reference Vergara, Ruiz-González, López de Luzuriaga and Gómez-Moliner2014). In addition, using the Wizard Genomic DNA Purification Kit (Promega Italia, Milan, Italy), we extracted genetic material from five tissue samples of otters found as roadkill, to use as controls.
Before DNA extraction, spraint samples were placed in a vacuum to evaporate the remaining ethanol. We extracted DNA with the QIAamp DNA Stool Mini Kit (Quiagen S.r.l., Milan, Italy) and performed genotyping using 11 polymorphic autosomal microsatellite loci, chosen among those reported in previous studies (Dallas & Piertney, Reference Dallas and Piertney1998; Dallas et al., Reference Dallas, Bacon, Carss, Conroy, Green and Jefferies1999; Huang et al., Reference Hung, Li and Lee2004, Reference Huang, Hsu, Lee and Li2005; Hájková et al., Reference Hájková, Zemanová, Bryja, Hájek, Roche, Tkadlec and Zima2006; Kalz et al., Reference Kalz, Jewgenow and Fickel2006; Bonesi et al., Reference Bonesi, Hale and Macdonald2013; Lerone et al., Reference Lerone, De Castro, Imperi, Marrese, Carranza, Scorpio and Loy2014; Vergara et al., Reference Vergara, Ruiz-González, López de Luzuriaga and Gómez-Moliner2014), based on expected polymorphisms and size of PCR fragments (Table 1). We split the 11 primer pairs into two multiplex PCRs and labelled them with different fluorescent dyes for distinguishing the amplified fragments. We used the Multiplex PCR Kit (Quiagen S.r.l., Milan, Italy) for PCR amplifications, following the manufacturer's recommendations, except for the final volume, which was reduced to 25 μl. We sequenced PCR amplification fragments using the commercial fragment analysis service offered by Macrogen (Seoul, South Korea) and analysed electropherograms using GeneMarker 2.2.0 (SoftGenetics, State College, USA; see Tremolada et al., Reference Tremolada, Smiroldo, Verduci, Gatti, Boggioni and Gianfranceschi2020 for further details).
Table 1 Number of samples successfully amplified (N S) among the genotyped samples of otter Lutra lutra spraint collected on the Ticino River, Italy, during 2016–2018. The table shows amplification success per locus, number (N A) and size of alleles, and observed and expected heterozygosis (H O and H E, respectively).

We analysed the mitochondrial DNA control region to verify the genetic strain and determine the occurrence of hybridization with a residual native population. We accessed sequences from European and Asian otters from GenBank (National Center for Biotechnology Information, Bethesda, USA; Table 2) to pinpoint a short region amplifiable in poor and degraded faecal mtDNA and capable of identifying the origin of the genotypes.
Table 2 Accession number and origin of European and Asian otter sequences downloaded from GenBank (National Center for Biotechnology Information, Bethesda, USA).

Using the primers OTTDL3- and OTTDH5- (Mucci et al., Reference Mucci, Arrendal, Ansorge, Bailey, Bodner, Delibes and Randi2010), we amplified mtDNA from seven of the 25 samples. The reaction was performed using the following protocol: 2 minutes at 94 °C, followed by 40 cycles of three steps (30 seconds at 94 °C, 30 seconds at 55 °C, and 30 seconds at 72 °C), before a final extension of 10 minutes at 72 °C. We carried out Sanger sequencing using the reverse primer OTTDH5. Amplicons were separated on an ABI 3130XL automated sequencer (Applied Biosystems, Waltham, USA) and visualized and corrected using Seqscape 5.0 (Applied Biosystems, Waltham, USA).
We also included a haplotype (GenBank accession no. AY860320) from an individual otter hosted in Caramanico Otter Centre, Italy, as it belonged to the same captive-bred group as the reintroduced individuals. We used the mtDNA extracted from tissues of an individual found as roadkill in the lower valley of the River Ticino in 2007 as a reference sample.
We aligned the results and sequences from GenBank in Bioedit 7.0 (Hall, Reference Hall1999) and created a neighbour joining phylogenetic tree (1,000 bootstrap resamples) in MEGA 7 (Kumar et al., Reference Kumar, Stecher and Tamura2016) to identify the maternal lineage of Italian spraint samples. We used a sequence of the hairy-nosed otter Lutra sumatrana (GenBank accession no. AY860320) to root the tree.
Population viability analysis
We performed the population viability analysis using Vortex 10 (Lacy & Pollak, Reference Lacy and Pollak2018), the most commonly used package for modelling reintroduced populations (Armstrong & Reynolds, Reference Armstrong, Reynolds, Ewen, Armstrong, Parker and Seddon2012) and guiding conservation initiatives (Hamilton & Moller, Reference Hamilton and Moller1995; Gaona et al., Reference Gaona, Ferreras and Delibes1998). Vortex is an individual-based, stochastic model that applies Monte Carlo simulations to model the dynamics of populations by stepping through a series of events that describe both deterministic and stochastic processes (Lacy, Reference Lacy1993). Simulations allow the probabilistic assessment of (1) population size and genetic variation of extant populations, (2) extinction risk at specified intervals, and (3) mean time to extinction of those simulated populations that became extinct.
We retrieved biological parameters from the available literature (Table 3; Larsson & Ebenhard, Reference Larsson and Ebenhard1994; Ansorge et al., Reference Ansorge, Schipke and Zinke1997; Ebenhard, Reference Ebenhard2000; Hauer et al., Reference Hauer, Ansorge and Zinke2002; Kruuk, Reference Kruuk2006; Kuhn & Jacques, Reference Kuhn and Jacques2011), and assessed carrying capacity based on otter density in southern Italy (Prigioni et al., Reference Prigioni, Remonti, Balestrieri, Sgrosso, Priore, Mucci and Randi2006b). To account for the uncertainty about the number of otter pairs released and the possibility of unintentional supplementation by otters escaped from two enclosures in the 2–3 years following the start of the reintroduction project, we set the population size (N 0) at three different values (1–3 pairs) for each scenario and simulated the adding of a pair of individuals per year at time t 0 + 1 and t 0 + 2 (Table 3). The risk of a catastrophe was set at 3%, assuming that exceptional floods, which may occur at intervals of c. 30–35 years (Cattaneo et al., Reference Cattaneo, Maione, Mignosa and Tomirotti2000), could primarily affect the survival of juveniles. For each scenario, we ran 500 simulations over a 100-year period, defining extinction as the point when one sex was eliminated.
Table 3 Modelling scenarios for assessing the viability of the otter population reintroduced on the River Ticino, with demographic input data for Vortex simulations.
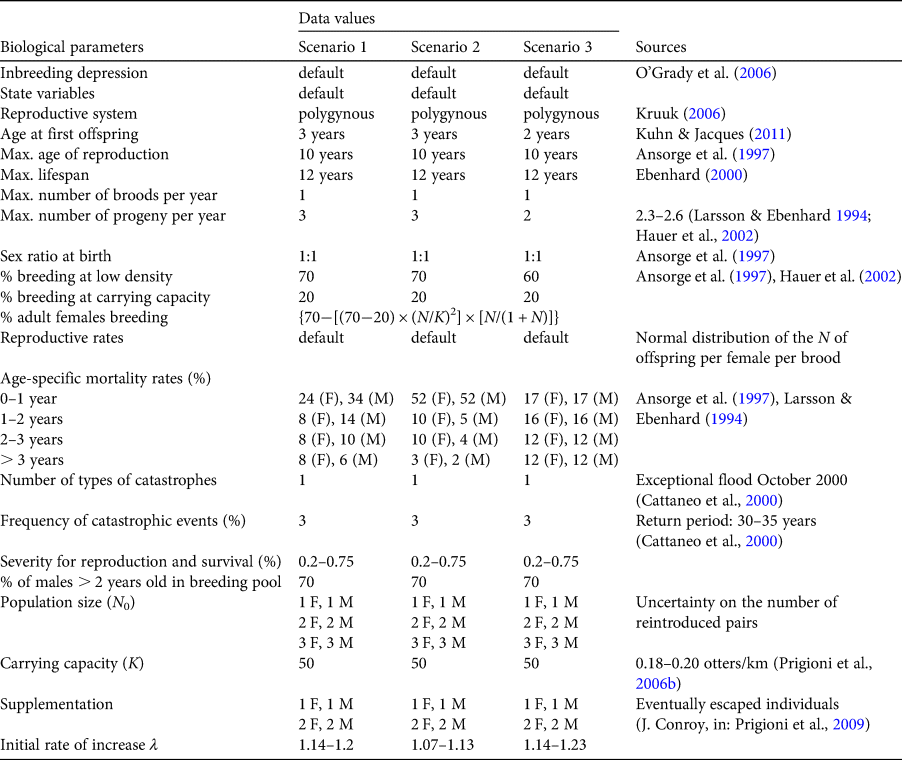
We conducted sensitivity tests for uncertain parameters, to examine the effects of a range of values for each parameter on the probability of otter survival. We tested inbreeding depression, quantified as the number of lethal equivalents (alleles that are lethal when homozygous or a collection of genes considered to be equivalent) per diploid individual (0.0–12.58, increments of 0.629), carrying capacity (K = 15–60, increments of 5), per cent frequency of catastrophic events (1–10%, increments of 1%) and severity of the latter for reproduction (0.1–1.0, increments of 0.1).
Results
Of the 25 faecal samples stored for genetic analyses (21 from monitoring in 2016–2017 and four from 2018), eight (32%) were successfully genotyped, allowing us to identify six individuals (five and one, respectively; Table 4), along a c. 36 km long stretch of the river (Fig. 1). The distance between faecal samples from the same individual was 4–11 km. Two loci were monomorphic and nine polymorphic (mean = 2.9 alleles per locus; maximum = 5 alleles for OT-04), for a total of 32 alleles. Expected heterozygosity (He) was 0.35 ± SE 0.07, and observed heterozygosity (Ho) was 0.41 ± SE 0.08.
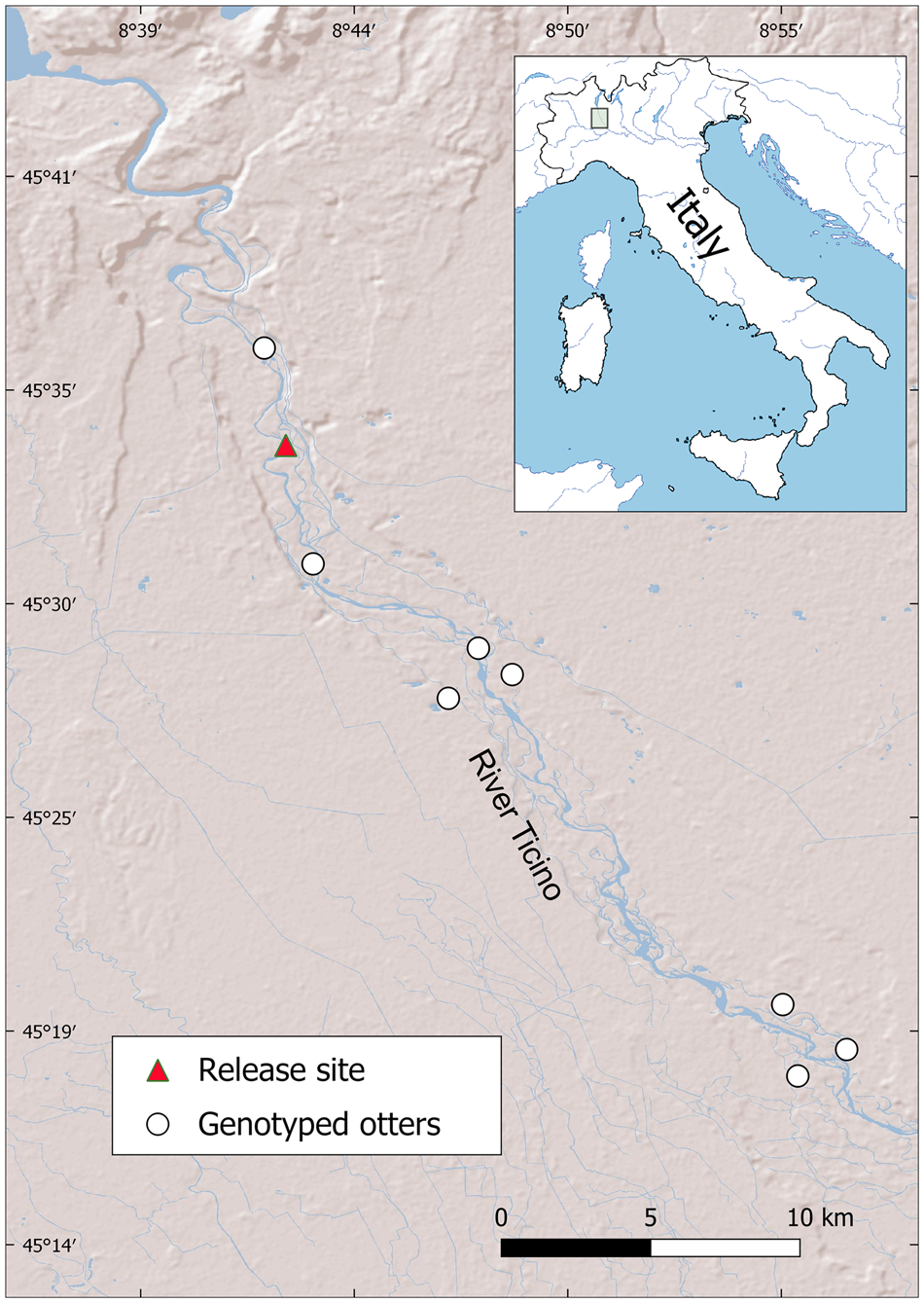
Fig. 1 Distribution of genotyped otter Lutra lutra spraints (2016–2018) on the River Ticino, Italy.
Table 4 Year of sampling, amplified loci and amplification success for the six otters genotyped.

We obtained a region of mtDNA 175 base pairs long for three of the seven analysed samples. All individuals shared an identical haplotype (Fig. 2) that was the same retrieved in the otter found as roadkill. All the Asian haplotypes were characterized by a transversion (A instead of G) at position 16.400 of the European lineage (Genebank accession no. MN122838). The same mutation was also found in the haplotype from the River Ticino.

Fig. 2 Neighbour joining tree showing the phylogenetic relationship between otters from the River Ticino (both non-invasive samples and one individual found dead on the road) and European, Chinese and Korean lineages.
Population size in 2016–2017 was consistent with population viability analysis models with N 0 = 2 and the supplementation of four individuals in the following 2 years (two per year; scenario 1), despite a high probability of extinction in the first decade post-release (Fig. 3). The reintroduction of such a small number of individuals would be insufficient to ensure the long-term survival of the population (P100 < 0.1; Fig. 3).
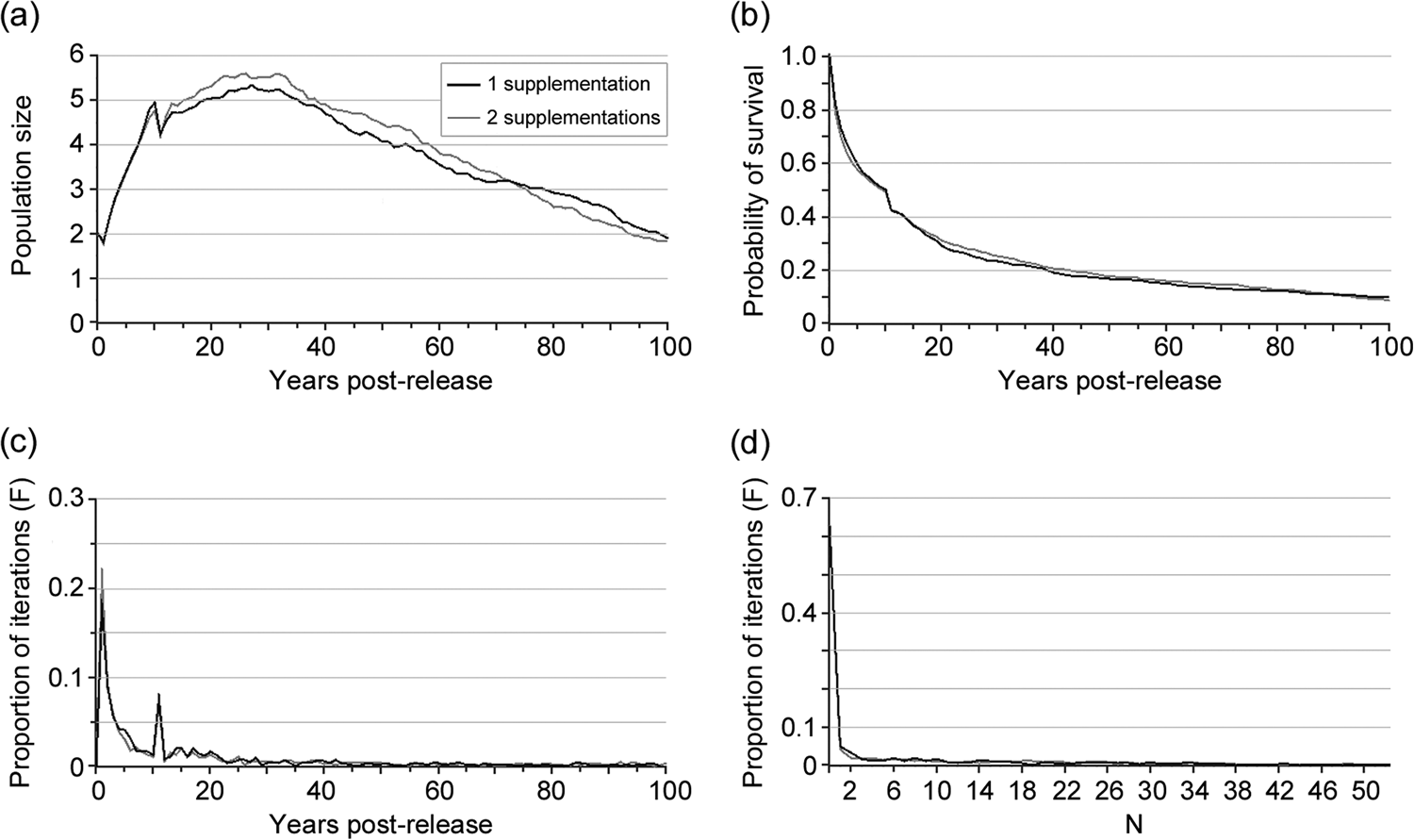
Fig. 3 Vortex simulations (scenario 1) for the reintroduced otter population, with initial population size N 0 = 2 and the supplementation of four individuals in the following years (two per year): (a) population trend, (b) probability of survival, (c) proportion of iterations (F) that lead to extinction, and (d) proportion of iterations that lead to population size = N, 21 years after the reintroduction.
For all scenarios, an initial phase of population growth, with a peak of 4–33 individuals (< K) 15–20 years after the reintroduction, was followed by a progressive decline leading to N 100 < N0 for all models, except for those of scenario 1 based on the concurrent release of 2–3 pairs (N 100 = 12 and 20 individuals, respectively). As expected, for most models the probability of extinction in the first decade post-release was high (c. 70%).
Sensitivity analyses indicated inbreeding depression as the main cause of variation in the probability of population survival (Fig. 4). For numbers of lethal equivalents below the default value proposed by Vortex it is an index of inbreeding depression used by Vortex, as added in the Methods (6.29), the probability of survival increased from 12 to 45%.

Fig. 4 Vortex sensitivity analysis, modelling the variation in the probability of survival of the reintroduced otter population for four uncertain parameters: (a) the number of lethal equivalents (increments of 0.629), (b) carrying capacity K (increments of 5), (c) per cent catastrophes (increments of 1%), and (d) severity for reproduction (increments of 0.1).
Discussion
Despite the small number of founder individuals and lack of post-release monitoring, an otter population has survived on the River Ticino since 1997. Signs of otter presence were recorded on a 7 km stretch of the river in 2010 (Prigioni & Balestrieri, Reference Prigioni and Balestrieri2011), and along 60 and 32 km in 2017 and 2018, respectively. Throughout the study period, the population comprised few individuals. Although the sample size and number of re-captures were insufficient to assess population size effectively, the number of individuals was considerably smaller than the predicted carrying capacity. The minimum number of individuals was consistent with that assessed by the most likely scenario in the population viability analysis, based on the information available: an initial population consisting of only one pair of otters, supplemented with a further pair per year in the following 2 years.
Faecal samples collected in 2016–2017 were stored for 1 year before extraction, whereas those found in 2018 were all analysed irrespective of how fresh they were. Despite this difference in the treatment of samples from different years, genotyping was reasonably successful, and our success rate of 32% was in the range of that reported in previous studies (e.g. 14%, Lanszki et al., Reference Lanszki, Hidas, Szentes, Révay, Lehoczky and Weiss2008; 21%, Ferrando et al., Reference Ferrando, Lecis, Domingo-Roura and Ponsà2008; 41%, Prigioni et al., Reference Prigioni, Remonti, Balestrieri, Sgrosso, Priore, Mucci and Randi2006b; 73%, Janssens et al., Reference Janssens, Fontaine, Michaux, Libois, De Kermabon, Defourny and Baret2008). Throughout the otter's European range, Ho is 0.35–0.69, and the mean number of alleles per locus is 4.9, ranging from 2.6 in southern Italy to 6.8 in Norway (Mucci et al., Reference Mucci, Arrendal, Ansorge, Bailey, Bodner, Delibes and Randi2010). Although these numbers cannot be directly compared, as four of our microsatellites differed from those used by Mucci et al. (Reference Mucci, Arrendal, Ansorge, Bailey, Bodner, Delibes and Randi2010), both the heterozygosity and polymorphism recorded in the Ticino population were as low as those reported for relict populations of southern Italy (Verduci, Reference Verduci2019), and consistent with the small founder population.
Although in the mid 1990s the EEP coordinator considered it unlikely that Asian otters had contributed to the programme's captive population (Vogt, Reference Vogt1995), mtDNA analysis confirmed the suspected B-line. Although otter signs are conspicuous, small isolated populations can remain undetected for several years (Gariano & Balestrieri, Reference Gariano and Balestrieri2018). Our findings do not support the hypothesis that a residual, autochthonous population persisted that may have been reinforced by the reintroduction programme.
Since reintroduction, the size of the otter population probably fluctuated randomly on the edge of extinction, between 2–3 and < 10 individuals, unable to spread throughout the aquatic habitats of the River Ticino valley. As the river has been judged suitable for otters by three different analyses (Prigioni, Reference Prigioni1995; Montanari & Boffino, Reference Montanari and Boffino2000; Prigioni & Balestrieri, Reference Prigioni and Balestrieri2011), the insufficient number of founders may be a major cause of low population growth.
All models in the population viability analysis suggest that the number of founders was also too few to ensure the long-term survival of the population; the simultaneous release of three pairs of otters would have increased population size and probability of persistence, although the risk of extinction in the short term would still have been high. Similarly, the extinction probability of a larger group of 10 otters in Uppland, Sweden, has been reported to be 49% within 50 years (Ebenhard, Reference Ebenhard2000). Such small groups also tend to lose half of the heterozygosity over 50 years, decreasing population fitness (Ebenhard, Reference Ebenhard2000; Reed & Frankham, Reference Reed and Frankham2003).
Inbreeding depression plays a major role in genetic stochasticity and increases extinction risk for most threatened mammals (O'Grady et al., Reference O'Grady, Broofe, Reed, Ballon, Tonkyn and Frankham2006). Consistently, according to population viability analysis, inbreeding depression is the factor that may contribute most to the variation in the extinction risk of the small populations on the River Ticino.
Considering that the genetic variability of the founder population was less than expected by EEP managers (Randi et al., Reference Randi, Davoli and Pierpaoli2001), there is reason to assume that the number of lethal equivalents was higher than the default value used by Vortex. Under such conditions, stochastic events such as reproduction failure or road mortality would be expected to drive this small isolated population to extinction.
There has been debate concerning a possible reinforcement of the reintroduced otter population (Balestrieri et al., Reference Balestrieri, Remonti, Prigioni and Angelici2016). Although B-line otters have also been reintroduced in the UK and the Netherlands (Duplaix & Savage, Reference Duplaix and Savage2018), the confirmed genetic composition of the Ticino population is a major hurdle to its reinforcement with wild-caught otters, for example from Austria, where otters are widespread and perceived as a threat by fish farmers (Kranz, Reference Kranz2000). On the other hand, although in the 2000s the population was completely isolated, recent expansion of otter populations in the French (Barthelemy & Arthur, Reference Barthelemy and Arthur2018), Austrian (Kranz, Reference Kranz2018) and Swiss Alps (Angst, Reference Angst2018) suggests that the River Ticino could be a potential route for future recolonization of northern Italy, and supports the need for further conservation actions. As a first step, any reinforcement project would require a new feasibility study and modelling of the release phase, to assess the number of individuals needed to achieve population viability in the long term.
The reintroduction project attempted on the River Ticino faced several of the 18 key problems described by Macdonald (Reference Macdonald, Hayward and Somers2009). In particular, it was not coordinated on a national level, the genetic origin of reintroduced animals was not considered, the number of individuals released was insufficient and there was no post-release monitoring. Despite this, in the last 2 decades the reintroduction has been considered a success, and a small population of otters persists on the river. Our findings highlight the need for long-term monitoring and viability analysis of reintroduced populations, as even positive trends in the first years post-release do not guarantee success in the longer term (Short, Reference Short2016).
Acknowledgements
The research was funded by Parco Lombardo della Valle del Ticino. We thank Adriano Bellani, Valentina Parco, Cristina Poma (Parco Lombardo della Valle del Ticino) and Paola Trovò (Parco del Ticino Piemontese) for logistic support; Giorgio Smiroldo, Alessandro Nessi, Elisa A. Bolzoni, Federico Poggio and Danila Rizzioli for help with fieldwork; and the Cambridge Conservation Initiative, Arcadia and Fondation Segré for funding the Endangered Landscape Programme ‘Restoring the Ticino River Basin Landscape. One River—Many Systems—One Landscape’.
Author contributions
Research conception: AB, PT; fieldwork: AB, PB; genetic analysis: LG, FV, EG, EC, NM, CM; data analysis: AB, LG, FV, PB; writing: AB, PT, NM.
Conflicts of interest
None.
Ethical standards
The research was non-intrusive and had the necessary approval and permits from the administration of Parco Lombardo della Valle del Ticino. The research conforms to the standards set out by the British Sociological Association and the Oryx guidelines on ethical standards.










