Introduction
Gastrointestinal infections constitute a common health problem worldwide, with pathogenic species of bacteria, viruses and parasites being the causative agents. Higher rates of these gastrointestinal parasitic infections have been more commonly reported in tropical and sub-tropical areas (Burd and Hinrichs, Reference Burd, Hinrichs and Leonard2016). Cryptosporidium spp. and Giardia duodenalis are among the parasites of zoonotic importance. These protozoan pathogens infect the gastrointestinal tract of a broad range of vertebrate hosts, including humans (Ryan et al., Reference Ryan, Feng, Fayer and Xiao2021). Their corresponding diseases, cryptosporidiosis and giardiasis are among the main causes of diarrhoea in both farm animals and humans (Einarsson et al., Reference Einarsson, Ma'ayeh and Svärd2016; Santin, Reference Santin2020). Among at least 48 recognized Cryptosporidium species, C. parvum is a major cause of diarrhoea in calves and lambs, resulting in significant economic losses (Roblin et al., Reference Roblin, Canniere, Barbier, Daandels, Dellevoet-Groenewegen, Pinto, Tsaousis, Leruste, Brainard, Hunter and Follet2023). Moreover, C. parvum and C. hominis are responsible for most cases of human cryptosporidiosis in both industrialized and non-industrialized countries (Xiao, Reference Xiao2010; Feng et al., Reference Feng, Ryan and Xiao2018). Giardia duodenalis is a multispecies complex comprising eight genetically distinct assemblages (genotypes; A-H), of which, A and B are the most reported zoonotic genotypes (Feng and Xiao, Reference Feng and Xiao2011; Cacciò et al., Reference Cacciò, Lalle and Svärd2018; Ryan et al., Reference Ryan, Feng, Fayer and Xiao2021). Human infections of Cryptosporidium and G. duodenalis can be asymptomatic or cause mild to severe gastrointestinal signs depending on species/genotypes/subtypes and/or the hosts' immune status.
Cryptosporidium spp. and G. duodenalis are transmitted via the faecal-oral route, following direct contact with infected hosts/environment or through consumption of contaminated water and foods. Livestock is an important reservoir and source of human infections by shedding of environmentally resilient Cryptosporidium oocysts and Giardia cysts. Both protozoans have previously been associated with both waterborne and foodborne outbreaks (Ryan et al., Reference Ryan, Hijjawi and Xiao2018; Ryan and Zahedi, Reference Ryan and Zahedi2019; Zahedi and Ryan, Reference Zahedi and Ryan2020). Outbreaks of cryptosporidiosis have been well documented in industrialised countries, including the USA, Ireland, UK, Finland, Germany, Sweden, Norway, South Korea, Australia and Canada (Karanis et al., Reference Karanis, Kourenti and Smith2007; Baldursson and Karanis, Reference Baldursson and Karanis2011; Zahedi and Ryan, Reference Zahedi and Ryan2020).Waterborne and foodborne outbreaks of giardiasis have also been reported in North America and Europe (Karanis et al., Reference Karanis, Kourenti and Smith2007; Baldursson and Karanis, Reference Baldursson and Karanis2011; Feng and Xiao, Reference Feng and Xiao2011). In comparison, data on outbreaks of gastrointestinal infections in non-industrialized countries have been poorly reported, primarily due to the absence of proper surveillance systems.
In Algeria, data on cryptosporidiosis and giardiasis are relatively poorly documented. Studies focusing on the prevalence and/or molecular characterisation of Cryptosporidium spp. are fragmentary and include cattle (Akam et al., Reference Akam, Lafri, Khelef, Kaidi, Bouchène, Cozma and Şuteu2007; Baroudi et al., Reference Baroudi, Khelef, Hakem, Abdelaziz, Chen, Lysen, Roellig and Xiao2017; Benhouda et al., Reference Benhouda, Hakem, Sannella, Benhouda and Cacciò2017; Sahraoui et al., Reference Sahraoui, Mammeri, Thomas, Chevillot, Polack, Vallée, Follet, Ain-Baaziz and Adjou2023), small ruminants (Baroudi et al., Reference Baroudi, Hakem, Adamu, Amer, Khelef, Adjou, Dahmani, Chen, Roellig, Feng and Xiao2018; Sahraoui et al., Reference Sahraoui, Thomas, Chevillot, Mammeri, Polack, Vallée, Follet, Ain-Baaziz and Adjou2019), equids (Laatamna et al., Reference Laatamna, Wagnerová, Sak, Květoňová, Aissi, Rost and Kváč2013, Reference Laatamna, Wagnerová, Sak, Květoňová, Xiao, Rost, McEvoy, Saadi, Aissi and Kváč2015), dromedary camels (Bouragba et al., Reference Bouragba, Laatamna, Cheddad, Baroudi, Houali and Hakem2020; Maxamhud et al., Reference Maxamhud, Reghaissia, Laatamna, Samari, Remdani, Gentekaki and Tsaousis2023), birds (Baroudi et al., Reference Baroudi, Khelef, Goucem, Adjou, Adamu, Zhang and Xiao2013; Laatamna et al., Reference Laatamna, Holubová, Sak and Kváč2017) and fish (Reghaissia et al., Reference Reghaissia, Maxamhud, Laatamna, Samari, Dahmane, Berima, Abdelli, Hakem, Baroudi and Tsaousis2022). Cryptosporidium parvum, C. hominis and C. felis were recently detected in Algerian HIV patients (Semmani et al., Reference Semmani, Costa, Achour, Cherchar, Ziane, Mouhajir, Villier, Hamoudi, Favennec and Razakandrainibe2023), revealing the presence of C. parvum IIa and IId subtype families, specifically IIaA14G2R1, IIaA15G2R1, IIaA16G2R1, IIaA20G1R1, IIaA21G1R1, IIdA16G1 and IIdA19G1 and C. hominis Ia and Ib subtype families, specifically IaA14, IaA22R2, IbA13G3 and IbA10G2. Giardia duodenalis has previously been detected in cattle (Baroudi et al., Reference Baroudi, Khelef, Hakem, Abdelaziz, Chen, Lysen, Roellig and Xiao2017), sheep (Sahraoui et al., Reference Sahraoui, Thomas, Chevillot, Mammeri, Polack, Vallée, Follet, Ain-Baaziz and Adjou2019; Benhassine et al., Reference Benhassine, Baroudi, Hakem, Thomas, Laatamna, Belkessa, Feng, Roellig, Chen, Adjou and Xiao2020) and humans (Rebih et al., Reference Rebih, Boutaiba, Aboualchamat, Souttou, Hakem and Al Nahhas2020; Belkessa et al., Reference Belkessa, Thomas-Lopez, Houali, Ghalmi and Stensvold2021a). Due to insufficient epidemiological data available from Algeria, transmission dynamics of Cryptosporidium and G. duodenalis among the Algerian populations, specifically those from rural areas, are poorly known.
The close proximity and interactions among humans, livestock and their environments play a significant role in disease transmission (Squire and Ryan, Reference Squire and Ryan2017). Livestock breeding is the main agricultural activity in the rural areas, particularly sheep breeding in the steppes and cattle and poultry farming in the high plateaus of Algeria. The absence of improved husbandry and management practices increases the risk for zoonotic transmission of both parasites. As such, understanding the transmission dynamics under a comprehensive framework requires a One Health approach.
The present study investigates the transmission dynamics of Cryptosporidium spp. and G. duodenalis between humans and animals from different households, and their environment in rural areas in Northeastern Algeria.
Materials and methods
Ethics statement
The ethics committee of the Laboratory of Exploration and Valorization of Steppic Ecosystems from the Faculty of Nature and Life Sciences, Ziane Achour University-Djelfa, Algeria, approved the collection of human samples in this study (001 EVES/FSNV/2021). The study protocol was explained to each family guardian (head of the family), and informed consent for collecting faecal samples was obtained.
Study area and sample collection
The present study was carried out between May and October 2022 in 5 rural areas; 4 in Guelma province and 1 in Souk Ahras province, both located in Northeastern Algeria (Fig. 1). Guelma is a highland region located in the Northeastern part of Algeria. It is characterized by diversified plant cover and sub-humid climate in the central and north parts, while in the south, the climate is semi-arid. In this region, there are valleys and dams, including Oued Seybouse. Souk Ahras is also a high plateau area located in the north-east and its climate is mainly semi-arid. This province has a dense hydrographic network, formed by a multitude of permanent and temporary watercourses such as Oued Medjerda, Oued Mellag and Oued Charef. In both provinces, livestock husbandry consists mainly of cattle, sheep and goat breeding. A total of 39 different households were included in this study, including 26 from Guelma and 13 from Souk Ahras. Each household was composed of 3–11 individuals, including both children and adults, at the time of sampling. These families lived in close proximity to various domestic animal species, including ruminants, birds, equids, dogs and cats. Moreover, stray dogs as well as wildlife, such as carnivores and boars are known to circulate in these areas. In both Guelma and Souk Ahras, households have access to nearby natural water sources.
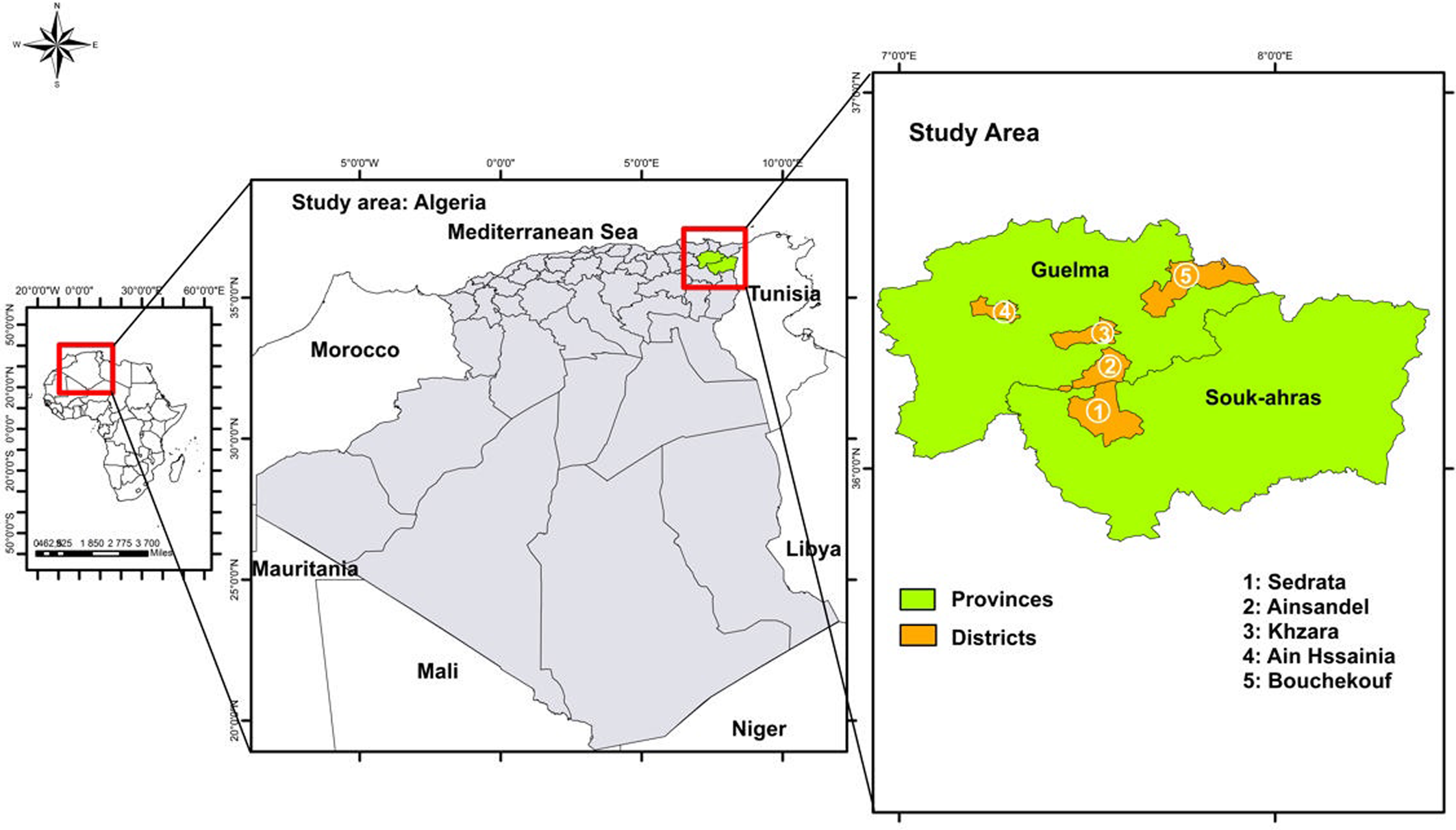
Figure 1. Map of Algeria, showing the 5 sampling regions in the 2 provinces: Guelma and Souk Ahras.
The inclusion criteria for participating in the study were living in rural areas and being clinically healthy (no gastrointestinal signs as well as no signs of any other disease). In this study, a total of 216 samples were collected. These consisted of human stools (n = 65), animal faeces (n = 135), water samples (n = 4) and soil samples (n = 12). More precisely, the collected animal samples consisted of cattle (n = 67), sheep (n = 56), chickens (n = 6), turkeys (n = 2), goats (n = 2), cat (n = 1) and horse (n = 1). Sterile tubes were distributed to members of the included households for stool collection. None of the sampled individuals showed any signs of diarrhoea at the time of sampling. The individuals sampled from each household ranged from 15% to 100% of the total residents within each household. In some households, no human samples, as well as no animal samples, were collected, only environmental. Data for sampled individuals, including the date of sampling, age, gender and their livestock, were obtained from the family guardian of each household. Faeces of animals were collected directly from the rectum or from the ground immediately after defecation. All faecal samples were preserved in DNA/RNA shield RNA solution at 4°C until molecular analysis.
Soil samples were collected close to the houses of the included families (n = 8) or, in some cases, from the nearest soil habitat to houses (n = 4). Water samples (n = 4) were sourced from drinking water, including 2 samples from natural water sources, one sample from a well and another from tap water.
DNA extraction and molecular analysis
The genomic DNA was extracted from 200 mg of both stool and soil samples using a PureLink™ Microbiome DNA Purification Kit (Invitrogen, Carlsbad, California, USA) according to the manufacturer's protocol. DNA from water samples was extracted using E.Z.N.A. Water DNA Kit (OMEGA, Stamford, CT, USA) according to the manufacturer's protocol. The extracted DNA was stored at −20°C until PCR analysis.
Detection of Cryptosporidium was performed using a nested PCR approach amplifying partial sequences of the small subunit rRNA (SSU) and 60-kDa glycoprotein (gp60) genes as previously described (Pinto et al., Reference Pinto, Ribeiro, Hoque, Hammouma, Leruste, Détriché, Canniere, Daandels, Dellevoet, Roemen, Bourgeois, Kváč, Follet and Tsaousis2021; Reghaissia et al., Reference Reghaissia, Maxamhud, Laatamna, Samari, Dahmane, Berima, Abdelli, Hakem, Baroudi and Tsaousis2022).
The process of Cryptosporidium spp. detection initially involved targeting and amplifying the SSU rRNA gene sequence for species/genotype determination. The primary reaction employed the primers CRY_SSU_F1 (5′-GATTAAGCCATGCATGTCTAA-3′) and CRY_SSU_R1 (5′-TTCCATGCTGGAGTATTCAAG-3′), resulting in a product size of 723 bp. Subsequently, for the second reaction, the forward primer CRY_SSU_F2 (5′-CAGTTATAGTTTACTTGATAATC-3′) and the reverse primer CRY_SSU_R2 (5′-CCTGCTTTAAGCACTCTAATTTTC-3′) was used, with an expected product size of approximately 631 bp (Lindergard et al., Reference Lindergard, Nydam, Wade, Schaaf and Mohammed2003). The primary PCR mixture, with a volume of 25 μL, contained 1 μL of template gDNA, 0.4 μm of each primer and 12.5 μL of 2 × PCRBIO Taq Mix Red. Amplification was carried out with an initial denaturation at 94°C for 2 min, followed by 24 cycles of denaturation at 94°C for 50 s, annealing at 53°C for 50 s and extension at 72°C for 1 min, concluding with a final extension step at 72°C for 10 min. For the secondary PCR reaction, 1 μL of the primary PCR product was utilized as a template, and the remaining mixture was prepared similarly to the primary reaction. In the second PCR condition, an initial denaturation was conducted at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 50 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min, concluding with a final extension step at 72°C for 10 min.
Furthermore, the amplification of the gp60 gene for Cryptosporidium parvun species subtyping was conducted via nested PCR using specific primers. In the primary reaction, the primers AL3531 (5′-ATAGTCTCCGCTGTATTC-3′) and AL3535 (5′-GGAAGGAACGATGTATCT-3′) were employed, and in the secondary reaction, the primers AL3532 (5′-TCCGCTGTATTCTCAGCC-3′) and AL3534 (5′-GCAGAGGAACCAGCATC-3′) were used (Pinto et al., Reference Pinto, Ribeiro, Hoque, Hammouma, Leruste, Détriché, Canniere, Daandels, Dellevoet, Roemen, Bourgeois, Kváč, Follet and Tsaousis2021). Both primary and secondary PCR mixtures contained 2 μL of gDNA or the primary PCR product, 0.2 μm of each primer and 15 μL of 2 × PCRBIO Taq Mix Red in a total volume of 30 μL. PCR conditions included an initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 45 s and extension at 72°C for 1 min, with a final extension step at 72°C for 7 min. The same PCR conditions were applied to both primary and secondary reactions.
DNA of positive samples was subsequently extracted and purified using the GeneJET Gel Extraction Kit (ThermoFisher Scientific, CA, USA).
Detection of G. duodenalis was performed by a probe-based quantitative PCR (qPCR) targeting a partial sequence (62 bp) of the SSU gene as previously described (Verweij et al., Reference Verweij, Blangé, Templeton, Schinkel, Brienen, Van Rooyen, Van Lieshout and Polderman2004; Hove et al., Reference Hove, Schuurman, Kooistra, Möller, Van Lieshout and Verweij2007; Maxamhud et al., Reference Maxamhud, Reghaissia, Laatamna, Samari, Remdani, Gentekaki and Tsaousis2023). Each reaction was composed of a 20 μL total volume, comprising 10 μL of 1× Luna Universal Probe qPCR Master Mix (New England Biolabs, Massachusetts, USA), 0.8 μL of 0.4 μM G. duodenalis-specific primers; Giardia-80F (5′-GACGGCTCAGGACAACGGTT-3′) and Giardia-127R (5́-TTGCCAGCGGTGTCCG-3′), 0.4 μL of 0.2 μm G. duodenalis-specific probe (FAM-5′-CCCGCGGCGGTCCCTGCTAG-3′-black hole quencher), 1 μL of 0.5 μg μL−1 bovine serum Albumin (BSA) (Promega Madison, WI, USA), 5 μL of nuclease-free H2O (Promega Madison, WI, USA) and 2 μL of extracted DNA. Amplification was conducted with QuantStudio™ three with PCR conditions as follows: an initial denaturation step at 95°C for 2 min, followed by 50 cycles of 95°C for 15 s, annealing at 57°C for 30 s and a final elongation step at 72°C for 30 s.
The determination of G. duodenalis assemblages was performed using nested PCRs targeting the triosephosphate isomerase (tpi), β-giardin (bg) and the glutamate dehydrogenase (gdh) genes as previously described (Cacciò et al., Reference Cacciò, De Giacomo and Pozio2002, Reference Cacciò, Beck, Lalle, Marinculic and Pozio2008; Sulaiman et al., Reference Sulaiman, Fayer, Bern, Gilman, Trout, Schantz, Das, Lal and Xiao2003). DNA of positive samples was subsequently extracted and purified using the GeneJET Gel Extraction Kit (ThermoFisher Scientific, CA, USA).
Sequencing, DNA cloning and phylogenetic analyses
Sequencing of PCR products from both Cryptosporidium spp. and G. duodenalis positive samples was performed bidirectionally at Eurofins laboratory (Cologne, Germany). Positive samples for either Cryptosporidium spp. or G. duodenalis that exhibited further evidence of mixed infections or non-specific binding/amplification were cloned as described by Betts et al. (Reference Betts, Gentekaki, Thomasz, Breakell, Carpenter and Tsaousis2018).
Obtained sequences were visualized and edited manually with the software SnapGene Viewer v.6.0.2 and ChromasPro v.2.1.10. Subsequently, the identity of obtained sequences was checked using BLAST search (www.ncbi.nlm.nih.gov/blast). Moreover, two distinct datasets were constructed, 1 for SSU rRNA gene sequences for Cryptosporidium spp. and another for only tpi gene sequences of G. duodenalis. Sequence analyses for bg and gdh genes were not included due to the poor quality of the obtained sequences. These sequence datasets were aligned using MAFFT v.7 (Katoh and Toh, Reference Katoh and Toh2010). Automated removal of ambiguous regions from sequence alignment was performed using trimAL v.1.4 (Capella-Gutiérrez et al., Reference Capella-Gutiérrez, Silla-Martínez and Gabaldón2009). Alignments consisted of 31 taxa (sequences) and 467 sites for G. duodenalis and 71 taxa and 1674 for Cryptosporidium. Maximum likelihood (ML) phylogenetic trees with bootstrap support were inferred by using IQTREE v.2.0.3 (Nguyen et al., Reference Nguyen, Schmidt, Von Haeseler and Minh2015) and MEGA7 v.7.0 (Kumar et al., Reference Kumar, Stecher and Tamura2016), and subsequently visualized using FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Furthermore, to assess the genetic diversity among multiple sequences generated in the present study, the genetic distance was calculated using Kimura2 parameter with gamma distribution as implemented in MEGA7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). This analysis determined the percentage of genetic variation between the sequences, enabling simultaneous comparisons across multiple sequences. Generated nucleotide sequences from the present study were deposited in the GenBank database under the following accession numbers: PP484686, PP484689, PP484691- PP484697, PP484699, PP484701 and PP484705 for C. bovis, PP484687 and PP484702-PP484704 for C. parvum, PP481965, PP481967- PP481968 and PP481972- PP481973 for C. parvum IIaA17G2R1, PP481966 for C. parvum IIaA16G2R1, PP481969 for C. parvum IIaA13G1R1, PP481970 for C. parvum IIA15G2R1, PP481971 for C. parvum IIdA16G1, PP481974 for C. parvum IIaA14G1R1, PP484685, PP484688, PP484698 and PP484700 for C. andersoni, PP484690 for C. ryanae and PP481975-PP481979, PP481981-PP481986 for G. duodenalis assemblage E and PP481980 for G. duodenalis assemblage B.
Results
The ages of the individuals involved in this study varied. Children aged 4–14 years (n = 23), adolescents aged 15–17 years (n = 3) and adults aged 22 to 48 years (n = 31) were included. Eight individuals had no age data recorded. Moreover, the number of females and males was 17 and 40, respectively. The remaining 8 human samples that were included had no gender data recorded.
At least one protozoan was identified in 62% (24/39) of households investigated, involving either animals, humans or both. Cryptosporidium spp. was detected in 41% (16/39) of the households, while G. duodenalis was detected in 56% (22/39) of the households included in this study. Both Cryptosporidium spp. and G. duodenalis were detected in 33% (13/39) of the households included (Table 1).
Table 1. Distribution of Cryptosporidium species and subtypes and Giardia intestinalis assemblages in animals, humans and environmental samples from examined households
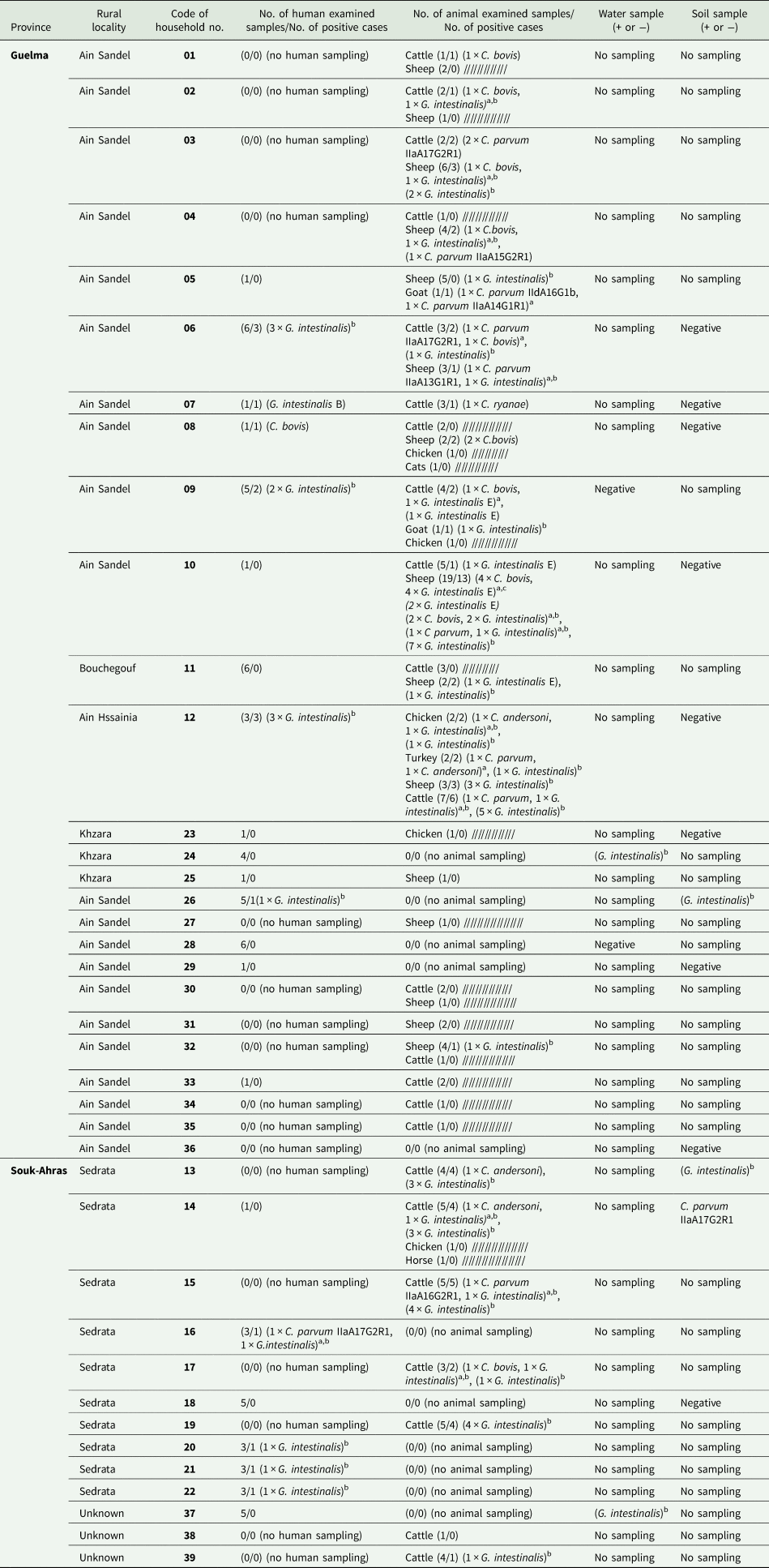
a Mixed infection.
b G. intestinalis positive samples with unsuccessful genotyping.
c Three different sheep samples showing mixed infection with C. bovis, and G. intestinalis assemblage E.
In total, out of the 135 animal samples screened, Cryptosporidium spp. was detected in 19% (26/135), which included 18% (12/67) of the sampled cattle, 20% (11/56) of the sampled sheep, 50% (1/2) of the sampled goats, 17% (1/6) of the sampled chickens and 50% (1/2) of the sampled turkeys. More precisely, C. bovis was present in 11 animals [42% of cattle (5/12) and 55% of sheep (6/11)], C. parvum in 10 animals [42% of cattle (5/12), 27% (3/11) of sheep, one goat and one turkey], C. andersoni in four animals [17% (2/12) of cattle, one chicken and one turkey] and C. ryanae (n = 1) in one cattle. One cattle from household N°6 and one turkey from household N°12 exhibited a mixed infection with C. parvum/C. bovis and C. parvum/C. andersoni, respectively. Samples that tested positive for C. parvum were subtyped as IIaA17G2R1 (n = 3) and IIaA16G2R1 (n = 1) in four different cattle, and IIaA15G2R1 (n = 1) and IIaA13G1R1 (n = 1) in two different sheep. In addition, one goat exhibited a mixed infection with two different C. parvum subtypes, namely IIdA16G1b and IIaA14G1R1. Three C. parvum samples, specifically one from sheep, one from turkey and one from cattle, could not be subtyped due to unsuccessful amplification of the gp60 gene. The sampled cat and horse were negative for both Cryptosporidium spp. and G. duodenalis.
Out of the 65 human samples examined in this study, 2 were positive for Cryptosporidium spp. This included one testing positive for C. bovis and the other for C. parvum (subtype IIaA17G2R1). These two samples were collected from two separate households. Neither participant showed signs of diarrhoea at the time of sampling. Moreover, in the household N° 8 (Table 1), C. bovis was detected in both humans and sheep.
Giardia duodenalis was detected by qPCR in 44% (59/135) of the animal samples, consisting of 45% (30/67) of cattle, 45% (25/56) of sheep, 50% (1/2) of goats, 33% (2/6) of chickens and 50% (1/2) of turkeys. Based on BLAST and phylogenetic analysis of tpi gene sequences (Fig. 2) ten animal samples, including seven from sheep and three from cattle, were successfully genotyped to assemblage E. In total, 17 animals exhibited mixed infections involving Cryptosporidium spp. and G. duodenalis. This included three cattle and eight sheep infected with C. bovis and G. duodenalis, two cattle and two sheep infected with C. parvum and G. duodenalis and one cattle and one chicken each infected with C. andersoni and G. duodenalis. Notably, among the sheep infected with C. bovis and G. duodenalis, four G. duodenalis samples were genotyped as assemblage E. These results were further confirmed with phylogenetic analyses (Fig. 2; Supplementary Fig. 1)

Figure 2. Phylogenetic relationship among Giardia duodenalis assemblages inferred based on partial sequences of the triosephosphate isomerase (tpi) gene using the maximum likelihood method.
Giardia duodenalis was detected in 14 samples of human origin, of which only 1 was successfully genotyped as assemblage B. One human sample exhibited a mixed infection with C. parvum (IIaA17G2R1) and G. duodenalis. Six of the G. duodenalis infected humans were children aged between 4 and 11 years, while 8 were adults aged between 27 and 84 years.
Giardia duodenalis was also detected in humans and animals residing within the same household. This included three different households, namely the households N° 6, 9 and 12 from Guelma province (Table 1). Additionally, G. duodenalis was identified in both humans and soil samples within the household N° 26 in Guelma.
Among the four analysed water samples, two natural sources were positive for G. duodenalis and have been successfully genotyped. These water-positive samples originated from two different households (N° 24, 37), and no human samples tested positive for G. duodenalis. Among the 12 analysed soil samples, 1 was positive for C. parvum (subtype IIaA17G2R1) and two for G. duodenalis which could not be successfully genotyped. One of the G. duodenalis positive soil samples was traced back to the household N°26 where a human sample also tested to be positive for G. duodenalis. The other G. duodenalis positive soil sample originated from the household N° 13 where three cattle were also positive to G. duodenalis. The soil sample that tested positive for C. parvum (subtype IIaA17G2R1) originated from a household, namely household N° 14, where no humans nor animals tested positive for this C. parvum subtype.
The samples that tested positive for G. duodenalis and were successfully genotyped originated from four distinct households, all located in Ain Sandel within the Guelma region. Among these, six sheep tested positive for assemblage E, exhibiting identical sequences, all belonging to the same household (N°10). Furthermore, two additional cattle samples, from a different household (N°9), also tested positive for assemblage E and shared identical sequences.
The sequence of C. andersoni was identical in 2 samples, 1 from a turkey and the other from a cattle. These samples originated from households N°12 and 14 in Ain Hssainia, Guelma province and Sedrata, Souk Ahras province, respectively (Table 1).
Additionally, a total of seven samples were positive for C. bovis, comprising 4 sheep, 2 cattle, and 1 human sample, shared identical sequences. These samples were collected from five distinct households (households N°1, 2, 4, 8 and 10), with three sheep originating from the household N°10. All these households were situated in Ain Sandel within the Guelma region. Furthermore, two other samples, both from cattle, also tested positive for C. bovis and exhibited identical sequences with each other but not with any other positive samples for C. bovis. One sample was obtained from the household N°17 in Ain Sandel, Guelma and the other from the household N°6 in Sedrata, Souk Ahras.
Discussion
This study took place in five rural areas, where traditional livestock breeding is the main activity. People in these areas live in close contact with their livestock (such as ruminants) as well as carnivorous pets (such as cats). Moreover, people rely on a local and traditional diet where cows' milk and meat are essential food sources. Hence, these communities are more exposed to the risk of transmission of infectious diseases. Due to these characteristics, this particular community was selected for this One Health study to gain insight into the transmission dynamics of 2 zoonotic protozoans: Cryptosporidium and G. duodenalis.
Among the animal species raised in the investigated households, cattle, sheep, goats, chickens and turkeys were infected with either Cryptosporidium or G. duodenalis, or both. This study reported the presence of G. duodenalis in domestic birds (chickens and turkeys), which could comprise a reservoir that contributes to environmental contamination in rural areas. In humans from the examined households, Cryptosporidium spp. and G. duodenalis were detected 3% and 21%, respectively. In the Algerians, human infections of both parasites have rarely been explored. A low prevalence (0.2%) of Cryptosporidium spp. was reported by Belkessa et al., (Reference Belkessa, Ait-Salem, Laatamna, Houali, Sönksen, Hakem, Bouchene, Ghalmi and Stensvold2021b), while another study reported an infection rate of 9.5% in HIV patients (Semmani et al. Reference Semmani, Costa, Achour, Cherchar, Ziane, Mouhajir, Villier, Hamoudi, Favennec and Razakandrainibe2023). Infection rates of G. duodenalis ranged of 0.3–67% (Benouis et al., Reference Benouis, Bekkouche and Benmansour2013; Hamaidi-Chergui et al., Reference Hamaidi-Chergui, Hamaidi, Benkhettar and Ould Mahieddine2013; Rebih et al., Reference Rebih, Boutaiba, Aboualchamat, Souttou, Hakem and Al Nahhas2020; Belkessa et al., Reference Belkessa, Thomas-Lopez, Houali, Ghalmi and Stensvold2021a, Reference Belkessa, Ait-Salem, Laatamna, Houali, Sönksen, Hakem, Bouchene, Ghalmi and Stensvold2021b). Various drivers can influence these prevalence variations such as sample size, detection methods, characteristics of persons involved in the study, and the study design.
Four Cryptosporidium species were detected in livestock raised in the investigated households, including C. andersoni, C. bovis, C. ryanae and C. parvum. Cryptosporidium bovis was the most detected species in cattle and sheep in the present study. In Algeria, C. bovis has been previously reported in pre-weaned calves (Baroudi et al., Reference Baroudi, Khelef, Hakem, Abdelaziz, Chen, Lysen, Roellig and Xiao2017; Benhouda et al., Reference Benhouda, Hakem, Sannella, Benhouda and Cacciò2017; Ouakli et al., Reference Ouakli, Belkhiri, de Lucio, Köster, Djoudi, Dadda, Khelef, Kaidi and Carmena2018; Sahraoui et al., Reference Sahraoui, Mammeri, Thomas, Chevillot, Polack, Vallée, Follet, Ain-Baaziz and Adjou2023) and in dromedary camels (Maxamhud et al., Reference Maxamhud, Reghaissia, Laatamna, Samari, Remdani, Gentekaki and Tsaousis2023). This species is well adapted to cattle, particularly in postweaning calves (Santin, Reference Santin2020), but it has been sporadically detected in other livestock, such as sheep (Ryan et al., Reference Ryan, Bath, Robertson, Read, Elliot, Mcinnes, Traub and Besier2005; Squire and Ryan, Reference Squire and Ryan2017). In the present study, one asymptomatic individual tested positive for C. bovis along with a sheep from the same household, suggesting possible direct transmission between the two. Human infections by C. bovis, specifically in cattle farm workers, have been reported in few cases, and therefore, its zoonotic importance seems to be minor (Khan et al., Reference Khan, Debnath, Pramanik, Xiao, Nozaki and Ganguly2010; Ng et al., Reference Ng, Eastwood, Walker, Durrheim, Massey, Porigneaux, Kemp, McKinnon, Laurie, Miller, Bramley and Ryan2012; Helmy et al., Reference Helmy, Krücken, Nöckler, von Samson-Himmelstjerna and Zessin2013). C. parvum was the second most detected species in livestock, including cattle, sheep, goats and turkey. C. parvum infects a broad host range and is the most common zoonotic Cryptosporidium species worldwide. The human sample that tested positive for C. parvum IIaA17G2R1, originated from a household where no animal sampling was done. Six different C. parvum subtypes that belonged to two zoonotic subtype families IIa and IId were identified in the present study. The subtype IIaA17G2R1 was identified in both cattle and humans with IIaA16G2R1 also detected in cattle. Cryptosporidium subtypes IIaA15G2R1 and IIaA13G1R1 were identified in sheep. In addition, one goat showed a mixed infection with IIaA14G1R1 and IIdA16G1b. In farm animals worldwide, subtype family IIa is commonly found in cattle, while subtype family IId is more commonly found in sheep and goats. However, subtype family IIa has also been reported in small ruminants (Xiao and Feng, Reference Xiao and Feng2017; Feng et al., Reference Feng, Ryan and Xiao2018). Cattle can contribute to the transmission of C. parvum subtypes of family IIa to sheep and goats (Feng et al., Reference Feng, Ryan and Xiao2018). Even though the infected human, who tested positive for C. parvum IIaA17G2R1, originated from a household where no animal and environmental samples were collected, it is important to note that cattle remain an important source of transmission for humans. In Algeria, the IIaA17G2R1 subtype has previously been detected in dromedary camels (Maxamhud et al., Reference Maxamhud, Reghaissia, Laatamna, Samari, Remdani, Gentekaki and Tsaousis2023) and fish (species Sparus aurata) (Reghaissia et al., Reference Reghaissia, Maxamhud, Laatamna, Samari, Dahmane, Berima, Abdelli, Hakem, Baroudi and Tsaousis2022). The present study reports, for the first time, the occurrence of IIaA17G2R1 in humans from Algeria. Moreover, the IIaA16G2R1 subtype has previously been detected in calves (Baroudi et al., Reference Baroudi, Khelef, Hakem, Abdelaziz, Chen, Lysen, Roellig and Xiao2017; Ouakli et al., Reference Ouakli, Belkhiri, de Lucio, Köster, Djoudi, Dadda, Khelef, Kaidi and Carmena2018; Sahraoui et al., Reference Sahraoui, Mammeri, Thomas, Chevillot, Polack, Vallée, Follet, Ain-Baaziz and Adjou2023), fish (Reghaissia et al., Reference Reghaissia, Maxamhud, Laatamna, Samari, Dahmane, Berima, Abdelli, Hakem, Baroudi and Tsaousis2022) as well as in HIV patients (Semmani et al., Reference Semmani, Costa, Achour, Cherchar, Ziane, Mouhajir, Villier, Hamoudi, Favennec and Razakandrainibe2023). The subtype IIaA15G2R1 that was identified in sheep in this study has also previously been reported in Algeria in calves (Sahraoui et al., Reference Sahraoui, Mammeri, Thomas, Chevillot, Polack, Vallée, Follet, Ain-Baaziz and Adjou2023) and dromedary camels (Maxamhud et al., Reference Maxamhud, Reghaissia, Laatamna, Samari, Remdani, Gentekaki and Tsaousis2023) as well as in HIV patients (Semmani et al., Reference Semmani, Costa, Achour, Cherchar, Ziane, Mouhajir, Villier, Hamoudi, Favennec and Razakandrainibe2023). The subtype IIaA15G2R1 is a common (mainly in cattle) and a hypertransmissible zoonotic subtype reported worldwide (Feng et al., Reference Feng, Ryan and Xiao2018) and herein we suggest that sheep could be an important source for human transmission and subsequent infection. The Cryptosporidium parvum subtype IIaA13G1R1 identified in sheep has not previously been reported in Algeria. Among the two examined goats, one exhibited a mixed infection with IIdA16G1b and IIaA14G1R1. Our results represented the detection of subtype families IIa and IId in goats from Algeria. The subtype IIaA14G1R1 has not been reported in Algeria in cattle or other livestock such as small ruminants. Other previous Algerian studies reported the occurrence of IIdA16G1 in lambs and calves (Sahraoui et al., Reference Sahraoui, Thomas, Chevillot, Mammeri, Polack, Vallée, Follet, Ain-Baaziz and Adjou2019, Reference Sahraoui, Mammeri, Thomas, Chevillot, Polack, Vallée, Follet, Ain-Baaziz and Adjou2023) and in HIV patients (Semmani et al., Reference Semmani, Costa, Achour, Cherchar, Ziane, Mouhajir, Villier, Hamoudi, Favennec and Razakandrainibe2023).
In the present study, C. andersoni was detected in cattle, chicken and turkey. This gastric species is predominant in juvenile and adult cattle, whereas it has been found less frequently in other livestock (Deng et al., Reference Deng, Li, Zhong, Gong, Cao, Song, Wang, Huang, Liu, Hu, Fu, He, Wang, Zhang, Wu and Peng2017; El-Alfy et al., Reference El-Alfy, Abu-Elwafa, Abbas, Al-Araby, Al-Kappany, Umeda and Nishikawa2019; Santin, Reference Santin2020). In Algeria, C. andersoni has been previously reported in calves (Baroudi et al., Reference Baroudi, Khelef, Hakem, Abdelaziz, Chen, Lysen, Roellig and Xiao2017). Mixed farming and close contact among cattle, sheep and domestic birds in the investigated rural areas could have contributed to the infection of non-specific hosts such as chickens and turkeys. Cryptosporidium andersoni is not considered a major zoonotic species, but it has been reported in humans in some countries (Santin, Reference Santin2020; Ryan et al., Reference Ryan, Feng, Fayer and Xiao2021). Additionally, C. ryanae was identified in one sample of cattle origin. This species is predominant in postweaned calves, but it has been found in small ruminants (Santin, Reference Santin2020). In Algeria, C. ryanae has previously been reported in preweaned calves (Benhouda et al., Reference Benhouda, Hakem, Sannella, Benhouda and Cacciò2017; Ouakli et al., Reference Ouakli, Belkhiri, de Lucio, Köster, Djoudi, Dadda, Khelef, Kaidi and Carmena2018). Cryptosporidium ryanae has no zoonotic impact and, to the best of our knowledge, has not been reported in humans (Ryan et al., Reference Ryan, Feng, Fayer and Xiao2021).
In the examined animals from the present study, positive G. duodenalis samples were successfully genotyped in cattle and sheep and identified as assemblage E. In Algeria, assemblage E has been reported from one study in lambs (Benhassine et al., Reference Benhassine, Baroudi, Hakem, Thomas, Laatamna, Belkessa, Feng, Roellig, Chen, Adjou and Xiao2020). This assemblage is common in hoofed animals such as ruminants and pigs (Feng and Xiao, Reference Feng and Xiao2011; Ryan and Zahedi, Reference Ryan and Zahedi2019). Only one sample was successfully identified as assemblage B in the examined humans. Assemblage B has been previously reported in Algerian children and adults (Belkessa et al., Reference Belkessa, Thomas-Lopez, Houali, Ghalmi and Stensvold2021a) but is commonly reported in humans (Ryan and Zahedi, Reference Ryan and Zahedi2019), in contrast to assemblage E (Fantinatti et al., Reference Fantinatti, Bello, Fernandes and Da-Cruz2016; Zahedi et al., Reference Zahedi, Field and Ryan2017). Different households showed the presence of G. duodenalis in humans and their livestock; however, despite our unsuccessful attempts to genotype the Giardia samples, this study emphasizes the possibility of infected livestock contributing to human infection and environmental contamination.
Among the four water samples screened for the presence of both parasites, twp natural water sources showed contamination by G. duodenalis. These contaminated samples originated from two different households, and the screened humans were negative for G. duodenalis and Cryptosporidium spp. Although livestock living close to these drinking water sources were not sampled, their contamination was likely associated with the presence of nearby infected animals.
Soil samples showed one positive for C. parvum (subtype IIaA17G2R1) and two for G. duodenalis. The soil sample tested positive for C. parvum IIaA17G2R1 and was collected in an area close to a household where cattle were infected with C. andersoni. However, as shown in the other examined households, cattle can act as an important reservoir and source for the environmental spread of this subtype. In the case of G. duodenalis, environmental soil contamination can be associated with faeces from infected livestock. It is important to note that the environmental contamination by Cryptosporidium oocysts and Giardia cysts can effectively contribute to human and livestock infections through environmental transmission. Waterborne outbreaks caused by Cryptosporidium spp. and G. duodenalis have been reported mainly in developed countries, while there is a lack of data on these outbreaks in developing countries (Karanis et al., Reference Karanis, Kourenti and Smith2007; Baldursson and Karanis, Reference Baldursson and Karanis2011). The present study provides preliminary data on the occurrence of C. parvum (subtype IIaA17G2R1) in soil and G. duodenalis in both soil and drinking water in Algeria. Epidemiological data on cryptosporidiosis and giardiasis waterborne outbreaks are poorly documented in developing countries, including those in the African continent. This is primarily due to a lack of technological and surveillance resources such as reliable diagnostic methods and effective reporting systems (Karanis et al., Reference Karanis, Kourenti and Smith2007; Baldursson and Karanis, Reference Baldursson and Karanis2011).
This study has some limitations, particularly concerning the imbalance of sampling and limited genotype/subtype-specific data, which did not allow for a clear understanding of the transmission dynamics. However, the role livestock and environment play in the possible transmission of human infections of Cryptosporidium spp. and G. duodenalis was demonstrated here. This study underscored the importance of adopting a One Health approach, particularly in rural settings where these parasites pose a significant challenge to human and animal health. This study did not only shed light on the genetic diversity of Cryptosporidium spp. across various hosts in Algeria, but also contributed to our growing knowledge of the molecular epidemiology of cryptosporidiosis and giardiasis in the country. Future comprehensive investigations are necessary in rural areas to enhance the understanding of transmission dynamics of these organisms. By emphasizing the relevance of the One Health concept as well as the transmission dynamics of these intestinal parasites between humans, animals and their shared environment, future effective control measures could be established.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024001616
Data availability statement
Generated nucleotide sequences from the present study were deposited in the GenBank database under the following accession numbers: PP484686, PP484689, PP484691-PP484697, PP484699, PP484701 and PP484705 for C. bovis, PP484687 and PP484702-PP484704 for C. parvum, PP481965, PP481967PP481968 and PP481972-PP481973 for C. parvum IIaA17G2R1, PP481966 for C. parvum IIaA16G2R1, PP481969 for C. parvum IIaA13G1R1, PP481970 for C. parvum IIA15G2R1, PP481971 for C. parvum IIdA16G1, PP481974 for C. parvum IIaA14G1R1, PP484685, PP484688, PP484698 and PP484700 for C. andersoni, PP484690 for C. ryanae and PP481975-PP481979, PP481981-PP481986 for G. duodenalis assemblage E and PP481980 for G. duodenalis assemblage B.
Acknowledgements
We thank members of the Tsaousis laboratory and the Algerian heads of households for assisting with sample collection and accommodating us during our visitations.
Author contributions
S. M.: Conceptualization, Data analysis, Manuscript drafting, Executing laboratory experiments, Review & Editing. N. R.: Conceptualization, Sampling, Manuscript drafting, Review & Editing. A. L.: Conceptualization, Project administration, Data analysis, Manuscript drafting, Review & Editing. A. T.: Conceptualization, Project administration, Supervising, Manuscript drafting, Review & Editing. E. G.: Data analysis, Project administration, Supervising, Manuscript drafting, Review & Editing.
Financial support
SM was supported by a Ph.D. studentship from the Global Challenges Doctoral Centre at the University of Kent.
Competing interests
The authors declare there are no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Ethical standards
The ethics committee of the Laboratory of Exploration and Valorization of Steppic Ecosystems from the Faculty of Nature and Life Sciences, Ziane Achour University-Djelfa, Algeria approved the collection of human samples in this study (001 EVES/FSNV/2021).