Introduction
Vector-borne haemosporidian parasites can negatively impact host fitness by mediating life-history trade-offs, such as trading investment in immune defence over investment in plumage coloration in response to infection (Hõrak et al., Reference Hõrak, Ots, Vellau, Spottiswoode and Møller2001; Delhaye et al., Reference Delhaye, Glaizot and Christe2018; Penha et al., Reference Penha, Rodrigues, Quaglia, Hoepers, Del-Claro and Soares2020). Furthermore, haemosporidian infections have been associated with avian mortality (Permin and Juhl, Reference Permin and Juhl2002; Atkinson and Samuel, Reference Atkinson and Samuel2010; Jia et al., Reference Jia, Huang, Valkiunas, Yang, Zheng, Pu, Zhang, Dong, Suo and Zhang2018), and with lower health status in birds (Himmel et al., Reference Himmel, Harl, Matt and Weissenböck2021). Haemosporidian parasites cause malaria and related diseases in wild and domesticated birds; these parasites are ecologically and evolutionarily diverse, with a worldwide distribution (Valkiūnas, Reference Valkiūnas2005; Perkins, Reference Perkins2014). Each haemosporidian genus is transmitted to the avian host by a different group of dipteran vectors: Plasmodium by mosquitoes (Culicidae) and Parahaemoproteus by biting midges (Ceratopogonidae; Santiago-Alarcon et al., Reference Santiago-Alarcon, Palinauskas and Schaefer2012). Because avian haemosporidian parasites are broadly distributed, common in avian populations, and easily detected in small blood samples, they provide an important and accessible model system for studying host–parasite interactions.
Within an avian community, host exposure to parasites may be influenced by the environment (e.g. climate), and life-history traits of the host species (Svensson-Coelho et al., Reference Svensson-Coelho, Ellis, Loiselle, Blake and Ricklefs2014; Canard et al., Reference Canard, Mouquet, Mouillot, Stanko, Miklisova and Gravel2015; Lutz et al., Reference Lutz, Hochachka, Engel, Bell, Tkach, Bates, Hackett and Weckstein2015; Clark and Clegg, Reference Clark and Clegg2017). Climate (particularly rainfall and temperature) may play an important role in parasite exposure through its influence on vector development and abundance (Loiseau et al., Reference Loiseau, Harrigan, Robert, Bowie, Thomassen, Smith and Sehgal2011; Gehman et al., Reference Gehman, Hall and Byers2018). For example, in central and west Africa, Plasmodium prevalence in the olive sunbird (Cyanomitra olivacea) was higher in locations with high temperatures (Sehgal et al., Reference Sehgal, Buermann, Harrigan, Bonneaud, Loiseau, Chasar, Sepil, Valkiunas, Iezhova, Saatchi and Smith2011). In community level studies, involving several avian host species, temperature also seems to be a good predictor of Plasmodium prevalence, such as in northeastern Brazil (Rodrigues et al., Reference Rodrigues, Felix, Pichorim, Moreira and Braga2021), and in the Spanish Iberian Peninsula (Illera et al., Reference Illera, López, García-Padilla and Moreno2017). However, Parahaemoproteus prevalence has shown contrasting results (associated with colder environments) in comparison with Plasmodium (Clark, Reference Clark2018; Clark et al., Reference Clark, Clegg, Sam, Goulding, Koane and Wells2018, Reference Clark, Drovetski and Voelker2020), which may be related to the different life histories of the primary vectors of Plasmodium and Parahaemoproteus parasites.
Host life-history traits may influence haemosporidian parasite prevalence, since these traits are associated with varying host exposure to vectors (Medeiros et al., Reference Medeiros, Hamer and Ricklefs2013; Svensson-Coelho et al., Reference Svensson-Coelho, Loiselle, Blake and Ricklefs2016). Nesting and foraging height, body size, habitat type, flocking, migratory behaviour (Møller and Erritzøe, Reference Møller and Erritzøe1998; Svensson-Coelho et al., Reference Svensson-Coelho, Loiselle, Blake and Ricklefs2016), and diet (González et al., Reference González, Matta, Ellis, Miller, Ricklefs and Gutiérrez2014; Turcotte et al., Reference Turcotte, Bélisle, Pelletier and Garant2018; Tchoumbou et al., Reference Tchoumbou, Mayi, Malange, Foncha, Kowo, Fru-cho, Tchuinkam, Awah-Ndukum, Dorazio, Nota Anong, Cornel and Sehgal2020) are all factors that may influence host exposure to vectors (Medeiros et al., Reference Medeiros, Hamer and Ricklefs2013; González et al., Reference González, Matta, Ellis, Miller, Ricklefs and Gutiérrez2014; Lutz et al., Reference Lutz, Hochachka, Engel, Bell, Tkach, Bates, Hackett and Weckstein2015). For example, single- or mixed-species flock participants tend to have a higher haemosporidian parasite prevalence because flocking hosts tend to attract more vectors or simply be in contact with more insects (Fecchio et al., Reference Fecchio, Lima, Svensson-Coelho, Marini and Ricklefs2013; Isaksson et al., Reference Isaksson, Sepil, Baramidze and Sheldon2013; Ellis et al., Reference Ellis, Medeiros, Collins, Sari, Coffey, Dickerson, Lugarini, Stratford, Henry, Merrill, Matthews, Hanson, Roberts, Joyce, Kunkel and Ricklefs2017), whereas birds foraging and nesting in the canopy and inhabiting closed habitats may have increased parasite prevalence due to a higher vector abundance in these forest strata (Garvin and Greiner, Reference Garvin and Greiner2003; Swanson and Adler, Reference Swanson and Adler2010; Laporta et al., Reference Laporta, Ramos, Ribeiro and Sallum2011; Swanson et al., Reference Swanson, Adler and Malmqvist2012; Ibañez-Justicia and Cianci, Reference Ibañez-Justicia and Cianci2015; Lutz et al., Reference Lutz, Hochachka, Engel, Bell, Tkach, Bates, Hackett and Weckstein2015). Host diet may also be an important factor in predicting haemosporidian prevalence, with insectivores harbouring higher prevalence, because of their closer contact with insects, which leads to an increased susceptibility to vectors (Braga et al., Reference Braga, Silveira, Belo and Valkiũnas2011; González et al., Reference González, Matta, Ellis, Miller, Ricklefs and Gutiérrez2014). Analyses of the influence of migration on haemosporidian prevalence have shown contrasting patterns; migratory host species have exhibited higher haemosporidian prevalence due to higher pathogen exposure (Ciloglu et al., Reference Ciloglu, Ergen, Inci, Dik, Duzlu, Onder, Yetismis, Bensch, Valkiūnas and Yildirim2020; Anjos et al., Reference Anjos, Chagas, Fecchio, Schunck, Costa-Nascimento, Monteiro, Mathias, Bell, Guimarães, Comiche, Valkiūnas and Kirchgatter2021; de Angeli Dutra et al., Reference de Angeli Dutra, Fecchio, Martins Braga and Poulin2021), but in other studies resident species have exhibited higher haemosporidian prevalence perhaps due to the increased predictability of hosts to vectors through space and time (Slowinski et al., Reference Slowinski, Fudickar, Hughes, Mettler, Gorbatenko, Spellman, Ketterson and Atwell2018; Soares et al., Reference Soares, Latta and Ricklefs2020). Haemosporidian parasite infection prevalence might also relate to host incubation period (Matthews et al., Reference Matthews, Ellis, Hanson, Roberts, Ricklefs and Collins2016), which is likely associated with avian life-history trade-offs between immune response and the duration of incubation (Ricklefs, Reference Ricklefs1992). Therefore, birds with longer incubation periods may have an adaptive advantage by having an increased length of time for B-cell maturation, conferring increased protection against infections (Ricklefs et al., Reference Ricklefs, Ellis, Medeiros and Svensson-Coelho2018).
Here, we investigated haemosporidian parasite prevalence in tanagers (Passeriformes: Thraupidae), the largest family of songbirds. Tanager species commonly occur from northern Mexico, through Central America, the Caribbean and South America, accounting for 12% of bird species in the Neotropical region (Parker et al., Reference Parker, Stotz, Fitzpatrick, Stotz, Fitzpatrick, Parker and Moskovits1996). Tanagers occupy several habitat types, ranging from rainforests to grasslands, with nearly all avian foraging niches being filled by members of the family (Burns et al., Reference Burns, Shultz, Title, Mason, Barker, Klicka, Lanyon and Lovette2014). Thraupidae currently includes 377 species placed in 15 subfamilies (Burns et al., Reference Burns, Unitt and Mason2016; Winkler et al., Reference Winkler, Billerman, Lovette, Billerman, Keeney, Rodewald and Schulenberg2020). Tanager species have a broad range of complex behaviours, habitat preferences, and morphological characteristics (Macedo et al., Reference Macedo, Manica and Dias2012; Manica and Marini, Reference Manica and Marini2012; Burns et al., Reference Burns, Shultz, Title, Mason, Barker, Klicka, Lanyon and Lovette2014; Nogueira et al., Reference Nogueira, Pope, Burke and Alves2014; Lima-Rezende and Caparroz, Reference Lima-Rezende and Caparroz2016; Beier et al., Reference Beier, Repenning, Da Silveira Pereira, Pereira and Fontana2017). Because of this impressive diversity, the accumulated knowledge on tanager ecology (Shultz and Burns, Reference Shultz and Burns2017), and the fact that they have been well sampled within the Neotropical region, make them a good model system for studying the effects of host life-history variation and environmental variation on haemosporidian prevalence. Despite recent advances in the study of haemosporidian prevalence of Neotropical birds (Fecchio et al., Reference Fecchio, Lima, Silveira, Braga and Marini2011, Reference Fecchio, Dias, Ferreira, Reyes, Dispoto, Weckstein, Bell, Tkach and Pinho2022; Sebaio et al., Reference Sebaio, Braga, Branquinho, Fecchio and Marini2012; de Angeli Dutra et al., Reference de Angeli Dutra, Fecchio, Martins Braga and Poulin2021; Ellis et al., Reference Ellis, Fecchio and Ricklefs2021), there is still a lack of information on the vulnerability of tanager species to haemosporidian parasites. Therefore, in this study we sought to understand the relationships among parasitism by haemosporidians, tanager life-history traits and environmental traits. More specifically, we tested whether haemosporidian parasite prevalence was related to species' nesting and foraging strata, habitat preference in terms of forest cover, participation in mixed-species flocks, diet, migratory behaviour, length of incubation period, environmental temperature regime, and annual precipitation.
Materials and methods
Data collection
We assembled haemosporidian screening data from 3569 individual birds belonging to 53 species in the family Thraupidae. Individuals were captured between 2007 and 2018 at 92 locations in 7 countries in the Neotropics, including Argentina (Soares et al., Reference Soares, Escudero, Penha and Ricklefs2016; Fecchio et al., Reference Fecchio, Bell, Pinheiro, Cueto, Gorosito, Lutz, Gaiotti, Paiva, França, Toledo-Lima, Tolentino, Pinho, Tkach, Fontana, Grande, Santillán, Caparroz, Roos, Bessa, Nogueira, Moura, Nolasco, Comiche, Kirchgartter, Guimarães, Dispoto, Marini, Weckstein, Batata-Filho and Collins2019a), Brazil (Lacorte et al., Reference Lacorte, Félix, Pinheiro, Chaves, Neto, Neves, Leite, Santos and Braga2013; Ferreira et al., Reference Ferreira, Rodrigues, Ellis, Leite, Borges and Braga2017; Fecchio et al., Reference Fecchio, Bell, Pinheiro, Cueto, Gorosito, Lutz, Gaiotti, Paiva, França, Toledo-Lima, Tolentino, Pinho, Tkach, Fontana, Grande, Santillán, Caparroz, Roos, Bessa, Nogueira, Moura, Nolasco, Comiche, Kirchgartter, Guimarães, Dispoto, Marini, Weckstein, Batata-Filho and Collins2019a, Reference Fecchio, Ribeiro, Ferreira, de Angeli Dutra, Tolesano-Pascoli, Alquezar, Khan, Pichorim, Moreira, Costa-Nascimento, Monteiro, Mathias, Guimarães, Simões, Braga, Kirchgatter and Dias2021; Lopes et al., Reference Lopes, Costa, Rodrigues, Braga, Pichorim and Moreira2020; Penha et al., Reference Penha, Rodrigues, Quaglia, Hoepers, Del-Claro and Soares2020; Rodrigues et al., Reference Rodrigues, Massara, Bailey, Pichorim, Moreira and Braga2020), Dominican Republic (Latta and Ricklefs, Reference Latta and Ricklefs2010; Soares et al., Reference Soares, Latta and Ricklefs2020), Ecuador (Svensson-Coelho et al., Reference Svensson-Coelho, Ellis, Loiselle, Blake and Ricklefs2014), Honduras (this study), Mexico (Fecchio et al., Reference Fecchio, Collins, Bell, García-Trejo, Sánchez-González, Dispoto, Rice and Weckstein2019b), and Nicaragua (this study).
Haemosporidian parasite analysis
To compare lineages identified by our nested PCR protocols to those in the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009), we aligned nucleotide sequences using the BIOEDIT v 7.2.0 program (Hall, Reference Hall1999) and verified sequence identities through a local BLAST against the MalAvi database. MalAvi is a database that groups and standardizes haemosporidian parasite lineages found in various hosts, allowing the study of host–parasite distributions, prevalence, and specializations (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009). Lineages identified using the protocol that amplified a longer mtDNA fragment (Ricklefs et al., Reference Ricklefs, Swanson, Fallon, Martínez-Abraín, Scheuerlein, Gray and Latta2005; Soares et al., Reference Soares, Escudero, Penha and Ricklefs2016, Reference Soares, Latta and Ricklefs2020) were successfully matched to known lineages in the MalAvi database only when the 2 fragments had 100% identical nucleotide sequences in their overlapping region (lineage names here are as in the MalAvi database). We calculated the prevalence of each Parahaemoproteus and Plasmodium lineage separately for every host species as the number of infected individuals divided by the total number of screened individuals (proportion of infected individuals). We treated Parahaemoproteus as a distinct genus from Haemoproteus (Haemoproteus) following recent phylogenetic advancements in the haemosporidian parasite phylogeny (Martinsen et al., Reference Martinsen, Perkins and Schall2008; Borner et al., Reference Borner, Pick, Thiede, Kolawole, Kingsley, Schulze, Cottontail, Wellinghausen, Schmidt-Chanasit, Bruchhaus and Burmester2016; Galen et al., Reference Galen, Borner, Martinsen, Schaer, Austin, West, Perkins and Galen2018).
Host phylogeny
We used the Thraupidae phylogeny from Burns et al. (Reference Burns, Shultz, Title, Mason, Barker, Klicka, Lanyon and Lovette2014), reconstructed with 6 molecular markers, which was the first comprehensive tanager phylogeny; Burns et al.'s (Reference Burns, Shultz, Title, Mason, Barker, Klicka, Lanyon and Lovette2014) phylogenetic hypothesis included genera not found in Jetz et al. (Reference Jetz, Thomas, Joy, Hartmann and Mooers2012). This phylogeny produced a highly comprehensive framework for studying macroevolutionary patterns among tanager taxa. We used ape (Paradis et al., Reference Paradis, Claude and Strimmer2004) to prune out species not found in our database from the tree.
Life-history traits and climate
We used the Handbook of the Birds of the World (Winkler et al.,, Reference Winkler, Billerman, Lovette, Billerman, Keeney, Rodewald and Schulenberg2020; https://birdsoftheworld.org/bow/home) to compile the following variables from the 53 tanager species: body size (average body length in centimetres); mixed-flock participation (participant [frequently or loosely join mixed-species flocks] and non-participant [rarely or does not join mixed-species flocks]); foraging height (ground [forages on or close to ground]; understory [forages in the midstory of the forest, understory, shrubs or small trees] and canopy [forages in tall trees, or in the canopy of forests]); migration status (migrant or permanent resident – complemented with data from Somenzari et al. (Reference Somenzari, do Amaral, Cueto, de Guaraldo, Jahn, Lima, Lima, Lugarini, Machado, Martinez, do Nascimento, Pacheco, Paludo, Prestes, Serafini, Silveira, de Sousa, de Sousa, de Souza, Telino-Júnior and Whitney2018) for species that occur in Brazil); and incubation period (average number of days laying to hatching). We also collected information on nest height, including low (0–1 m; on or close to the ground), middle (1–5 m; in shrubs, small trees, understory or mid canopy) and high (>5 m or tall trees and upper canopies). We used the data available in Olson and Owens (Reference Olson and Owens2005) to categorize foraging ecology including plant-eating (herbivore: fruits, seeds, leaves and other plant parts), animal-eating (carnivore: arthropods, spiders or others) or a generalist diet (omnivore). We used the data available in the Global Habitat Heterogeneity database (Tuanmu and Jetz, Reference Tuanmu and Jetz2015) as a proxy for habitat type (denoted by the variable name ‘forest cover’ hereafter). We used occurrence data from eBird (https://ebird.org/data/download) and the extract function from the raster package (Hijmans, Reference Hijmans2021), and then averaged the GHH) (Global Habitat Heterogeneity) values for each species across its distribution. Higher GHH indicates more forested habitats, whereas a lower GHH indicates open habitats. Lastly, we extracted all 19 climatic variables for the capture sites of all individuals (our 92 different capture sites) from WorldClim 2 (Fick and Hijmans, Reference Fick and Hijmans2017). For each host species, we averaged climatic values from all sites where a given species was captured. Since we could not determine age and sex of all individuals from every species, we did not include these 2 variables in our models. We then performed a principal components analysis to reduce the dimensionality of the climatic variables (summary statistics can be found in Supplementary Table 2 and Fig. 2). The first and second components together explained 68.6% of the variation and were used as our climatic variables (hereinafter climate PC1 and PC2). PC1 was primarily related to temperature and was positively associated with variables such as mean annual temperature, minimum temperature of coldest month, mean temperature of driest quarter and mean temperature of the coldest quarter, whereas PC2 was negatively associated with precipitation variables, such as annual precipitation, precipitation of the driest month, precipitation of the driest quarter, and positively associated with precipitation seasonality.
Statistical analysis
Using the host phylogeny, we created 2 different phylogenetic generalized least-squares models to test the hypothesis that parasite prevalence is predicted by host-related parameters and climate. For each model we used parasite prevalence (proportion of infected individuals) as the response variable, one for Parahaemoproteus, and another for Plasmodium. We only considered species with 5 or more captured individuals for analysis with these models (see Supplementary Tables 3–5 and Fig. 3 for a more conservative analysis including species with 10 or more captured individuals). We used the following explanatory variables: climate PC1, climate PC2, body size, mixed-species flock participation, incubation period, migration, nest height, foraging height, forest cover and diet. All numerical variables were standardized using the scale function from R, to remove unwanted variation in the scale among variables. Before including all variables, we tested for multicollinearity using the variance inflation factor (VIF) calculated by using the VIF function from the regclass package (James et al., Reference James, Witten, Hastie and Tibshirani2014; Petrie, Reference Petrie2020). We used a conservative threshold of 2 for the values of GVIF(1/2df) as a sign of multicollinearity. We found no collinear predictors based on this approach, and therefore all variables were included in the analysis. We tested model convergence with the Ornstein–Uhlenbeck (OU) and Brownian motion (BM) evolutionary models using Akaike information criterion (AIC) values. We then selected the best models using an information-theoretic approach (Burnham and Anderson, Reference Burnham and Anderson2002) with the dredge function in the MuMIn package (Barton, Reference Barton2019). When wi (weight) of the best model was below 0.80, we used model averaging in the model.avg function in the MuMIn package to calculate the model-averaged estimates, following the protocol described by Burnham et al. (Reference Burnham, Anderson and Huyvaert2011). We used root mean square error (RMSE) to validate each model, considering RMSE closest to zero as models with a good fit (Norberg et al., Reference Norberg, Abrego, Blanchet, Adler, Anderson, Anttila, Araújo, Dallas, Dunson, Elith, Foster, Fox, Franklin, Godsoe, Guisan, O'Hara, Hill, Holt, Hui, Husby, Kålås, Lehikoinen, Luoto, Mod, Newell, Renner, Roslin, Soininen, Thuiller, Vanhatalo, Warton, White, Zimmermann, Gravel and Ovaskainen2019; Tobler et al., Reference Tobler, Kéry, Hui, Guillera-Arroita, Knaus and Sattler2019; Snell Taylor et al., Reference Snell Taylor, di Cecco and Hurlbert2021). We assessed the importance of the explanatory variables by evaluating their estimates, unconditional standard errors and 95% confidence intervals (CIs) in the averaged model. Since foraging, nest height and diet have 3 different levels, we used the relevel function to change the reference level of each categorical variable to rerun the model and check for a specific pattern of statistical significance. Therefore, we only considered foraging, nest height and diet as significant if a level was different from all other levels. We plotted all significant variables using the ggplot2 (Wickham, Reference Wickham2016) package. All values are presented as mean ± s.d., unless otherwise noted.
Results
Haemosporidian parasites
From a total of 3569 screened individuals, we found 1469 birds infected with haemosporidian parasites (41% overall prevalence). We found 88 different Plasmodium lineages and 64 Parahaemoproteus lineages, with Parahaemoproteus prevalence marginally higher (16%) than Plasmodium (13%).
Host life-history traits and climatic variables
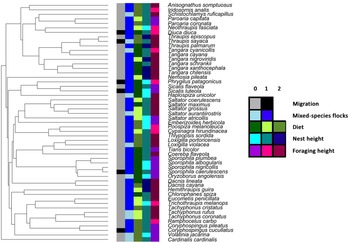
We found that most of the tanager species were mixed-species flock participants (79%), non-migratory (88%), middle-forest strata nest builders (60%) and canopy foragers (52%, Fig. 1). Host main diet was well-balanced among the species, with 39% herbivores, 34% omnivores and 27% carnivores (Fig. 1; Supplementary Table 2). Average body size was 14.7 ± 3.2 cm, and incubation period was 13.2 ± 1.0 days. Most of the host species also occurred in more open habitats (Fig. 2).

Fig. 1. Summary data for categorical life-history variables mapped onto the tips of the trimmed tanager phylogeny, showing as follows: migration (0 – resident; 1 – migrant); mixed-species flocking (0 – non-participant; 1 – participant); diet (0 – plant; 1 – animal; 2 – omnivore); nest height (0 – low; 1 – middle; 2 – high) and foraging height (0 – ground; 1 – understory; 2 – canopy). The colour keys for each category of life-history variables can be seen on the right inset.

Fig. 2. Forest cover histogram multiplied by 0.0001, showing that most species inhabit more open habitats (forest cover closer to zero indicates less forest cover). Forest cover data retrieved from Global Habitat Heterogeneity – dissimilarity index (https://www.earthenv.org/texture), which contains imagery from Moderate Resolution Imaging Spectroradiometer (MODIS) with pixel values collected from satellite images.
Prevalence models
The best models for Parahaemoproteus prevalence are presented in Table 1 (RMSE = 0.81). We found higher Parahaemoproteus prevalence among tanager species inhabiting areas with less forest cover (Table 2; Fig. 3), and with longer incubation periods (Table 2, Fig. 4). We also found a positive relationship between Parahaemoproteus prevalence and incubation period in a more conservative analytical approach (Supplementary Tables 4 and 6).

Fig. 3. Parahaemoproteus prevalence in relation to forest cover (in logarithmic scale) at host specimen collection locations. Points represent the observed values of Parahaemoproteus prevalence, and the black line is the fitted curve to the values with the standard error (shaded area).

Fig. 4. Parahaemoproteus prevalence in relation to the incubation period (average number of days). Points represent the observed values for the model incorporating Parahaemoproteus prevalence, and the black line is the fitted curve to the values with the standard error (shaded area).
Table 1. Model selection results of Parahaemoproteus and Plasmodium prevalence (response variables) and the following explanatory variables: climate PC1, climate PC2, body size, mixed-species flock participation, incubation, migration, nest height, foraging height, forest cover and diet

Variables included in each model are shown together with the models' degrees of freedom (df), AICc score, delta AIC and weight (wi). We only show the models with AIC scores lower than 4 for Parahaemoproteus and Plasmodium. Results for all 53 sampled tanager species in total. Model comparison using OU (Parahaemoproteus model AIC = −62.78; Plasmodium model AIC = −85.83) and BM (Parahaemoproteus model AIC = −35.95; Plasmodium model AIC = −56.92), indicated OU as the best in all our models.
Table 2. Model-averaged estimates, standard errors and 95% CIs for variables in the model using Parahaemoproteus prevalence as the response variable

Significant variables are marked with asterisks. Results for all 53 sampled tanager species in total.
a Reference level for the categorical variables: diet (animal), foraging height (canopy), migration (migrant), nest height (high) and mixed-species flock participation (non-participant).
The best models of Plasmodium prevalence are shown in Table 1 (RMSE = 0.30). Tanager species without migratory behaviour, with omnivorous or animal-derived diet, mixed-species flocking behaviour (Table 3; Fig. 5) and inhabiting areas with lower forest cover (Table 3; Fig. 6) had higher Plasmodium prevalence. We found that mixed-species flock participants had a higher Plasmodium prevalence in a more conservative analytical approach (Supplementary Tables 4 and 6).

Fig. 5. Observed values of Plasmodium prevalence in relation to migration status (left) and diet (middle) and mixed-species flock participation (right). Letters indicate statistical difference in prevalence among hosts with different levels of migration status, diet and mixed-species flock participation, meaning that tanager species that migrate, have a plant-derived diet, and do not join mixed-species flocks have lower Plasmodium prevalence in comparison with tanager species that are resident, an omnivorous or animal-derived diet, and participate in mixed-species flocks, respectively.

Fig. 6. Observed values of Plasmodium prevalence in relation to the forest cover. Points represent the values by the model of Plasmodium prevalence in relation to forest cover, and the black line is the fitted curve to the values with the standard error (shaded area).
Table 3. Model-averaged estimates, standard errors and 95% CIs of variables in the model using Plasmodium prevalence as the response variable

Significant variables are marked with an asterisk. Results are for all 53 sampled tanager species in total.
a Reference level for the categorical variables: diet (animal), foraging height (canopy), migration (migrant), nest height (high) and mixed-species flock participation (non-participant).
b Changing the reference level to diet (omnivore): diet (plant): −0.21 ± 0.03 (−0.28, −0.15) and diet (animal): −0.03 ± 0.04 (−0.12, 0.05). Changing the reference level to diet (plant): diet (omnivore): 0.21 ± 0.03 (0.15, 0.28) and diet (animal): 0.18 ± 0.02 (0.13, 0.24).
c Changing the reference level to foraging height (ground): foraging height (understory): 0.01 ± 0.05 (−0.09, 0.11) and foraging height (canopy): −0.08 ± 0.04 (−0.16, 0.00). Changing the reference level to foraging height (understory): foraging height (ground): −0.03 ± 0.05 (−0.14, 0.08) and foraging height (canopy): −0.08 ± 0.04 (−0.18, 0.00).
Discussion
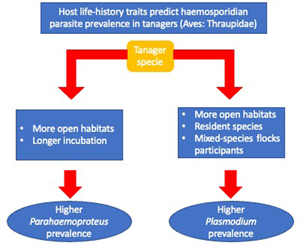
Overall, we found an association between haemosporidian parasite prevalence and tanagers' life-history traits. Specifically, we found that higher Parahaemoproteus prevalence was associated with birds occurring in habitats with lower forest cover (more open habitats), and among birds with longer incubation periods. We also found that Plasmodium prevalence was more often associated with birds without migratory behaviour, mixed-species flock participation, with an omnivorous or animal-derived diet and inhabiting less-forested habitats.
We found, first, that Parahaemoproteus and Plasmodium prevalence was higher in tanager species inhabiting locations with lower forest cover (more open habitats). Habitat type may be an important predictor of haemosporidian parasite prevalence because it may affect the probability of individual birds being exposed to vectors. Haemosporidian parasite vectors are common in nature and have shown some level of host specificity (Martínez-De La Puente et al., Reference Martínez-De La Puente, Martínez, Rivero-De Aguilar, Herrero and Merino2011a; Bobeva et al., Reference Bobeva, Zehtindjiev, Ilieva, Dimitrov, Mathis and Bensch2015; Tomás et al., Reference Tomás, Pereira da Fonseca, Valkenburg and Rebelo2021) and these vectors may change their feeding preferences according to the environmental conditions (Santiago-Alarcon et al., Reference Santiago-Alarcon, Palinauskas and Schaefer2012). The abundance and prevalence of biting midges can vary with altitude and across and habitat types (open vs closed) (Möhlmann et al., Reference Möhlmann, Wennergren, Tälle, Favia, Damiani, Bracchetti, Takken and Koenraadt2018), which may explain increased probability of infecting tanagers across our sampling locations. Furthermore, vectors of Plasmodium have been found to prefer pasture and more open areas in southeastern Brazil (Ferreira et al., Reference Ferreira, Rodrigues, Ellis, Leite, Borges and Braga2017). However, previous studies have reported contrasting results relating haemosporidian prevalence to habitat type, either showing higher parasite prevalence in open (Reinoso-Pérez et al., Reference Reinoso-Pérez, Canales-Delgadillo, Chapa-Vargas and Riego-Ruiz2016; Ferreira et al., Reference Ferreira, Rodrigues, Ellis, Leite, Borges and Braga2017) or in closed habitats (Lutz et al., Reference Lutz, Hochachka, Engel, Bell, Tkach, Bates, Hackett and Weckstein2015). Our results suggest that tanager species inhabiting places with less forest cover may be more exposed and therefore have an increased likelihood of encountering vectors carrying Parahaemoproteus and Plasmodium parasites, but future studies should identify these vectors as well as how their differences may vary across habitat types.
Second, we found that tanager species with a longer incubation period had higher Parahaemoproteus prevalence; this was the opposite of what we expected. These results were reinforced by findings using more conservative models (n > 10 individuals per species) for Parahaemoproteus prevalence. A longer incubation period is believed to allow for enhanced development of the immune system (Ricklefs, Reference Ricklefs1992), with higher B-cell maturation, thus conferring better defence against infections (Ricklefs et al., Reference Ricklefs, Ellis, Medeiros and Svensson-Coelho2018). However, based on our findings we hypothesize that tanager species facing higher selective pressure from Parahaemoproteus parasites may trade investing in reproduction over immunity, producing a weaker immune response to fight-off parasites. This is supported by other studies; for example, Palacios and Martin (Reference Palacios and Martin2006) found that a longer incubation period does not enhance cellular immune response in several passerine bird species. Alternatively, longer incubation periods may increase the chances of attracting vectors of haemosporidian parasites (biting midges for Parahaemoproteus; mosquitoes for Plasmodium) to incubating adults and their nestlings (Skutch, Reference Skutch1945; Santiago-Alarcon et al., Reference Santiago-Alarcon, Palinauskas and Schaefer2012) that may lead to more frequent or more efficient parasite infection during this period. Therefore, we also hypothesize that birds with longer incubation periods suffer increased susceptibility to Parahaemoproteus vectors among individuals or may attract mosquitoes (Plasmodium) more often.
Third, we found that birds joining mixed-species flocks, either frequently or rarely, had higher Plasmodium prevalence. Mixed-species flocks are thought to benefit participants through increased foraging success or increased surveillance against potential predators (Zou et al., Reference Zou, Jones, Colorado, Jiang, Lee, Martínez, Sieving, Zhang, Zhang and Goodale2018). In the Neotropics, birds often associate with mixed-species flocks after the breeding season to gain potential benefits (Kajiki et al., Reference Kajiki, Montaño-Centellas, Mangini, Colorado and Fanjul2018). However, like González et al. (Reference González, Matta, Ellis, Miller, Ricklefs and Gutiérrez2014), we show a positive relationship between flock participation and an increase in the probability of infection by Plasmodium parasites. This may be explained by (a) increasing visual or olfactory cues within mixed-species flocks that are in-turn associated with vector attraction (Díez-Fernández et al., Reference Díez-Fernández, Martínez-de la Puente, Gangoso, López, Soriguer, Martín and Figuerola2020), or (b) individual birds covering a larger spatial area within flocks resulting in an increased possibility of mosquito encounters (Van Houtan et al., Reference Van Houtan, Pimm, Bierregaard, Lovejoy and Stouffer2006).
Fourth, contrary to our original expectations, we found that resident tanager species had a higher Plasmodium prevalence compared to migratory tanager species. During migration movements, birds might be more exposed to vectors and, hence, present an increased likelihood of haemosporidian parasite infection (de Angeli Dutra et al., Reference de Angeli Dutra, Fecchio, Martins Braga and Poulin2021). However, non-migrating birds may become more predictably located in space and time, thus increasing their chances of encountering infected vectors year-around. For example, migratory passerines were seldom infected with haemosporidian parasites compared to resident birds in the Dominican Republic (Soares et al., Reference Soares, Latta and Ricklefs2020). Therefore, our results suggest 2 non-mutually exclusive hypotheses: (a) vectors may have a clear preference and be specialized in resident species, or (b) by encountering sedentary species more often, these species are more likely infected than migratory tanagers. However, because only 12% of our sampled species were migratory (mostly partially migratory), our results should be interpreted with caution; more studies are needed with a larger sample of the family Thraupidae. Furthermore, it is important to emphasize that haemosporidian prevalence, in a particular avian host species, could be an oversimplification as infection probability across individuals' hosts could still depend on other spatial and temporal variables not measured here (e.g. water body availability, bird breeding season), as well as individual development.
Finally, we found that omnivores and tanager species with a more animal-derived diet have a higher Plasmodium prevalence compared to those species feeding solely on plant materials. Feeding behaviour is crucial for bird survival, but costs may incur if foraging increases chances of encountering predators (Kelleher et al., Reference Kelleher, Hunnick and Sheriff2021), or vectors of haemosporidian parasites (Fecchio et al., Reference Fecchio, Dias, Ferreira, Reyes, Dispoto, Weckstein, Bell, Tkach and Pinho2022). In fact, our results suggest that birds seeking insects (animal-derived diets and omnivores) may face more encounters with infected vectors with haemosporidian parasites (Ribeiro et al., Reference Ribeiro, Sebaio, Branquinho, Marini, Vago and Braga2005). Furthermore, our results may also indicate that birds with a plant-derived diet have decreased infection chances simply because they have fewer encounters with insects considering that this is not their main feeding resource.
In summary, we found patterns of infection prevalence suggesting that parasitism by haemosporidians is related to a variety of tanager life-history traits. These findings for the host family Thraupidae highlight the difficulty in determining what factors affect parasite prevalence in birds. We suggest 2 non-mutually exclusive approaches to further clarify these relationships and to reveal whether reduced immune response and/or variability in exposure to vectors influences the infection susceptibility of hosts: (1) determining haemosporidian parasite prevalence within relevant vector species in relation to the habitats of avian hosts, and (2) analysing energy trade-offs between immunity and incubation period.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022001469.
Data availability
The authors comply with data availability criterion. Data used in this paper were provided along with R scripts and all datasets used for our analysis.
Acknowledgements
We thank countless ornithologists and field assistants who helped collect the blood samples used in this study. The curators and collections managers from the following museums loaned new samples used in this study: Academy of Natural Sciences of Drexel University and Yale Peabody Museum of Natural History. Also, we thank Dr Regina Macedo from Universidade de Brasília, Brasília, DF, Brazil, and Dr Diego Gill from the Spanish National Research Council for their support to R. D. A. and G. T.-P. Finally, G. T.-P. thanks Daniela de Angeli Dutra for all the help during lab analysis.
Author contributions
V. A. P. and L. T. M. designed research. V. A. P. performed the research, analysed the data and wrote the manuscript. All authors have provided data, and contributed to data analysis, writing and reviewing the manuscript.
Financial support
We acknowledge the governmental agencies that provided all permits necessary for collection and exportation of tissue samples. This study was funded in part by the U.S. National Science Foundation (DEB-1503804) to J. D. W. V. A. P. acknowledges Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the scholarship provided during the study. K. D.-C. and E. M. B. are grateful for a fellowship from Conselho Nacional de Ciência e Tecnologia (CNPq). R. E. R. is grateful for generous support from the U.S. National Science Foundation and the National Geographic Society.
Conflict of interest
The authors declare no conflicts of interest.
Ethical standards
All fieldwork was legally permitted in the above countries and was also approved by relevant Institutional Animal Care and Use Committees. Molecular protocols, primers used and PCR protocols, can be found in Bell et al. (Reference Bell, Weckstein, Fecchio and Tkach2015), Lacorte et al. (Reference Lacorte, Félix, Pinheiro, Chaves, Neto, Neves, Leite, Santos and Braga2013), Latta and Ricklefs (Reference Latta and Ricklefs2010), Lopes et al. (Reference Lopes, Costa, Rodrigues, Braga, Pichorim and Moreira2020), Rodrigues et al. (Reference Rodrigues, Massara, Bailey, Pichorim, Moreira and Braga2020), Svensson-Coelho et al. (Reference Svensson-Coelho, Ellis, Loiselle, Blake and Ricklefs2014), Soares et al. (Reference Soares, Escudero, Penha and Ricklefs2016, Reference Soares, Latta and Ricklefs2020) and Penha et al. (Reference Penha, Rodrigues, Quaglia, Hoepers, Del-Claro and Soares2020); Supplementary Tables 1, 2 and Fig. 1.