Introduction
The accepted view of the gastrointestinal (GI) tract is an open tube from the mouth to the anus that has evolved to control and interact with the external environment. The host GI tract is the largest immunological organ in the body; involving mechanisms to identify, control and eliminate dangerous pathogens present, while at the same time minimizing inappropriate inflammatory responses against benign antigens such as commensal bacteria and diet. In addition, the mammalian host GI tract also provides a favourable ecosystem with a stable temperature of approximately 37°C and a continual influx of nutrients that permit the growth of a complex microbiota. While numerically bacteria are the largest inhabitants of the GI tract, the GI tract has a number of diverse macro- and microbiotic communities which include viruses, fungi, protozoans and of particular interest for this review intestinal-dwelling parasitic nematodes.
Almost every mucosal surface on a mammal's body is heavily colonized with a variety of bacteria. Colonization begins at birth and initiates a variety of complex processes that lead to the establishment of a truly symbiotic relationship between the microbiota and host. We are only beginning to understand the numerous roles the microbiota plays in human health and the development, progression and resolution of disease. The gut microbiota (bacteria that inhabit the GI tract) has been the focus of intense research for the last few decades and is known to contribute to immunity, host metabolism and behaviour. The microbiota is known to mediate these functions by both direct (cell-to-cell) and indirect (bacterial-derived by-products) mechanisms which can involve interactions with the mammalian host immune cells during development (Elahi et al., Reference Elahi, Ertelt, Kinder, Jiang, Zhang, Xin, Chaturvedi, Strong, Qualls, Steinbrecher, Kalfa, Shaaban and Way2013; Gensollen et al., Reference Gensollen, Iyer, Kasper and Blumberg2016), hormone and behaviour signalling in the endocrine system (Lyte and Bailey, Reference Lyte and Bailey1997; Diaz Heijtz et al., Reference Diaz Heijtz, Wang, Anuar, Qian, Björkholm, Samuelsson, Hibberd, Forssberg and Pettersson2011), influencing disease progression of autoimmune disease such as inflammatory bowel disease (IBD), (Halfvarson et al., Reference Halfvarson, Brislawn, Lamendella, Vázquez-Baeza, Walters, Bramer, D'Amato, Bonfiglio, McDonald, Gonzalez, McClure, Dunklebarger, Knight and Jansson2017) or type 1 diabetes (Markle et al., Reference Markle, Frank, Mortin-Toth, Robertson, Feazel, Rolle-Kampczyk, von Bergen, McCoy, Macpherson and Danska2013), as well as affecting drug metabolism and efficacy (Zimmermann et al., Reference Zimmermann, Zimmermann-Kogadeeva, Wegmann and Goodman2019). From the time of birth, there is a known bidirectional interplay between the mammalian host and microbiota that is influenced by the composition, in terms of both diversity and abundance, of the gut microbiota which impacts host health (Dominguez-Bello et al., Reference Dominguez-Bello, Costello, Contreras, Magris, Hidalgo, Fierer and Knight2010).
While gut microbiota coevolves with the host immune system to influence host health, key factors that influence this process include enhanced sanitation and antibiotic usage, which are different in low/middle-income countries compared to high-income countries. Epidemiological evidence indicates that in high-income countries, there is low to no incidence of enteric parasite infections, but there has been a steady rise in autoimmune and allergic-related conditions, like asthma, eczema, diabetes and IBD in humans (Bach, Reference Bach2002). Improved diagnostic tools to identify the incidence and prevalence of disease have likely contributed to the rise in autoimmune and allergic-related conditions observed over time; however, it is also well documented that impeding parasite exposure (and the subsequent effects of parasitic infections) in early life results in a disruption of the normal immune development and leads to a greater predisposition to autoimmune and allergic diseases in life (Strachan, Reference Strachan1989; Bach, Reference Bach2002). This theory is called the ‘hygiene hypothesis’ (Strachan, Reference Strachan1989; Bach, Reference Bach2002), and has been further refined to the ‘microbial diversity’ hypothesis (also known as the ‘old friends’ hypothesis or ‘microflora hypothesis’) which proposes that enhanced sanitation and hygiene in high-income countries has resulted in a reduction of microbial exposure and/or gut microbiota composition in early life, that alters immune development and causes a predisposition to allergic diseases (Wold, Reference Wold1998; Rook, Reference Rook2009; Scudellari, Reference Scudellari2017). Therefore, the means by which the gut microbiota can impact human health is highly complex and varied, and there is growing evidence of epidemiological, microbiological and immunological data that support these hypotheses. However, there tends to be a focus on research that examines the bidirectional relationship between the microbiota and host. The presence of enteric multicellular parasites adds an additional level of complexity, and increases our understanding of the pervasive importance of the gut microbiota, and how these relationships impact mammalian host health and disease. Here we will focus on the interaction between the gut microbiota and enteric parasites, specifically whipworm infections by Trichuris and its dependency on the gut microbiota for propagation, persistent infection and survival in the mammalian gut.
The gut microbiota
The gut microbiota is a highly dynamic community of >1000 different bacterial species primarily comprised of phyla belonging to Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria and Verrucomicrobia (Arumugam et al., Reference Arumugam, Raes, Pelletier, Le Paslier, Yamada, Mende, Fernandes, Tap, Bruls, Batto, Bertalan, Borruel, Casellas, Fernandez, Gautier, Hansen, Hattori, Hayashi, Kleerebezem, Kurokawa, Leclerc, Levenez, Manichanh, Nielsen, Nielsen, Pons, Poulain, Qin, Sicheritz-Ponten, Tims, Torrents, Ugarte, Zoetendal, Wang, Guarner, Pedersen, de Vos, Brunak, Doré, Antolín, Artiguenave, Blottiere, Almeida, Brechot, Cara, Chervaux, Cultrone, Delorme, Denariaz, Dervyn, Foerstner, Friss, van de Guchte, Guedon, Haimet, Huber, van Hylckama-Vlieg, Jamet, Juste, Kaci, Knol, Lakhdari, Layec, Le Roux, Maguin, Mérieux, Melo Minardi, M'rini, Muller, Oozeer, Parkhill, Renault, Rescigno, Sanchez, Sunagawa, Torrejon, Turner, Vandemeulebrouck, Varela, Winogradsky, Zeller, Weissenbach, Ehrlich and Bork2011). In return for the host providing a protective, nutrient-rich niche for the gut microbiota to inhabit, some of these bacteria perform essential functions for host health including the synthesis of vitamins (such as vitamin K, Gustafsson, Reference Gustafsson1959), the degradation of complex nutrients (Bäckhed et al., Reference Bäckhed, Ley, Sonnenburg, Peterson and Gordon2005), limiting pathogen invasion (Stecher et al., Reference Stecher, Macpherson, Hapfelmeier, Kremer, Stallmach and Hardt2005) and contributing to immune system development (Stappenbeck et al., Reference Stappenbeck, Hooper and Gordon2002; Flint et al., Reference Flint, Scott, Louis and Duncan2012). During the first three years of life, there is a succession of colonization events, for humans, this is marked by events such as a diet change from milk to solid foods that promotes the development of a stable gut microbial composition that is able to contribute to host health and readily adapt to lifestyle changes over a life time (Wu et al., Reference Wu, Chen, Hoffmann, Bittinger, Chen, Keilbaugh, Bewtra, Knights, Walters, Knight, Sinha, Gilroy, Gupt, Baldassano, Nessel, Li, Bushman and Lewis2011). Antibiotic usage is the most robust and quick-acting factor that can alter microbial diversity, closely followed by microbial changes induced by changes to the host diet (Jakobsson et al., Reference Jakobsson, Jernberg, Andersson, Sjölund-Karlsson, Jansson and Engstrand2010; David et al., Reference David, Maurice, Carmody, Gootenberg, Button, Wolfe, Devlin, Varma, Fischbach, Biddinger, Dutton and Turnbaugh2013). An imbalance in the microbial community (referred to as a state of dysbiosis) can have a profound impact on increasing host susceptibility to disease and it is typically associated with an increase of facultative anaerobic bacteria (Shin et al., Reference Shin, Whon and Bae2015; Rizzatti et al., Reference Rizzatti, Lopetuso, Gibiino, Binda and Gasbarrini2017).
Technological advancements in the ability to decipher the composition and function of the microbiota, including targeted culturing techniques for recalcitrant strains, culture-independent techniques including 16S rRNA amplicon sequencing and whole-genome sequencing coupled with metabolomics and computational analysis have transformed our understanding of how important the gut microbiota is in host health (Browne et al., Reference Browne, Forster, Anonye, Kumar, Neville, Stares, Goulding and Lawley2016). Bacterial composition of the microbiota, in particular microbial diversity, is crucial, as it increases the likelihood of containing species that are capable of degrading complex carbohydrates into microbial fermentation products being present in the GI tract. The gut microbiota is dominated by obligate anaerobes, specifically the phyla Firmicutes (Gram-positive) and Bacteroides (Gram-negative). Members of these phyla are characterized by encoding a large variety of enzymes that hydrolyse complex dietary and host carbohydrates that provide benefit to the host (Flint et al., Reference Flint, Scott, Louis and Duncan2012; El Kaoutari et al., Reference El Kaoutari, Armougom, Gordon, Raoult and Henrissat2013). For example, short-chain fatty acids (SCFAs) are small organic carboxylic acids (with chain lengths up to six carbons), of which acetate (C2), propionate (C3) and butyrate (C4) are the most abundant metabolites produced from anaerobic fermentation of dietary fibre and resistant starch. There is growing evidence that SCFA function as metabolic moieties in crosstalk between the host and gut microbiota to exert immunomodulatory activity upon the host. Namely these metabolites have also been identified as an energy supply for colonocytes, regulation of T regulatory (Treg) cell populations in the mucosal immune system and a potential role for treatment of autoimmune diseases (Cummings et al., Reference Cummings, Pomare, Branch, Naylor and Macfarlane1987; Furusawa et al., Reference Furusawa, Obata, Fukuda, Endo, Nakato, Takahashi, Nakanishi, Uetake, Kato, Kato, Takahashi, Fukuda, Murakami, Miyauchi, Hino, Atarashi, Onawa, Fujimura, Lockett, Clarke, Topping, Tomita, Hori, Ohara, Morita, Koseki, Kikuchi, Honda, Hase and Ohno2013; Gill et al., Reference Gill, van Zelm, Muir and Gibson2018).
Germ-free animal models and Trichuris
Germ-free animals do not contain any detectable microorganisms, and are commonly used as a reductionist approach to investigating the relationship between the host and single (or known multiple-gnotobiotic) bacterial species (Smith et al., Reference Smith, McCoy and Macpherson2007; Flint et al., Reference Flint, Scott, Louis and Duncan2012). Recently these approaches have been used to triangulate the relationships between the host, its microbiota and intestinal multicellular organisms (such as helminths).
In the late 19th century, Louis Pasteur was the first to postulate that bacteria residing in the GI tract formed a relationship with the host that was critical for survival (Pasteur, Reference Pasteur1885). This theory was perceived to be true, until a decade later it was disproved by raising germ-free guinea-pigs, and demonstrating life was possible without bacteria (Nuttal and Thierfelder, Reference Nuttal and Thierfelder1895). However, throughout the early 20th century, scientists continued to pursue the understanding of the relationship between the host and the bacteria that inhabit the GI tract. Using a variety of animal models and human subjects, researchers found that bacteria must be confined to mucosal surfaces for health (including areas such as the intestinal lumen, nasal cavity and aspects of the reproductive system), and a breach of this containment resulted in severe sepsis and/or death (Cushing and Livingood, Reference Cushing and Livingood1900; Reyniers, Reference Reyniers1959). Today, germ-free mice are the most common germ-free model used due to the number of immune-comprised models available, and their ability to breed frequently with large litters in a single cage. Germ-free mice are born and raised in sterile flexible-film isolators maintained under positive pressure to the external environment, and receive sterile chow, bedding, water and HEPA-filtered sterile air to limit exposure to live microbial antigens (Smith et al., Reference Smith, McCoy and Macpherson2007). Interaction and manipulation of the gut microbiota by enteric worms as a potent mechanism to regulate the host immune system and worm persistence are emerging as a topic of study where the use of germ-free models can provide us insight into this intricate and complex relationship. For intestinal parasitic infections, such as Trichuris, there is a critical dependency on a living host for propagation and survival, making germ-free models a means to provide mechanistic insight into the relationship between bacteria and worm infection (Hayes et al., Reference Hayes, Bancroft, Goldrick, Portsmouth, Roberts and Grencis2010; Leung et al., Reference Leung, Budischak, Chung The, Hansen, Bowcutt, Neill, Shellman, Loke and Graham2018; White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018).
Helminth-microbe interactions
Microbial cues to induce Trichuris muris infection
Trichuris trichiura is one of the most prevalent soil-transmitted parasites, and infects approximately 500 million people worldwide (World Health Organization, 2020). Although whipworm infections rarely cause mortality in humans, individuals with a high worm burden can suffer from severe morbidities including diarrhoea/abdominal pain, malnutrition and infected children often have impaired growth/physical development that impacts their health status for life (World Health Organization, 2020). Bacterial diversity and density vary along the GI tract, with the densest microbial population residing within the caecum and colon of both mice and humans, reaching upwards of 100 trillion bacteria. Intestinal whipworms also inhabit this microbial-dense niche, and it is highly likely that these worms and the surrounding microbiota interact with each other (either directly or indirectly), as well as their mammalian host. An overview of how Trichuris interacts with the mammalian host, in particular the host immune system during acute and/or chronic infection, and the details of the Trichuris life cycle is presented in this special issue by Mair et al. (Reference Mair, Else and Forman2021).
To decipher how bacteria in the human GI tract interacts with T. trichiura, researchers use mouse models as the host, and infect mice with the murine-specific T. muris whipworm. Trichuris muris infection follows the same strict oral-faecal route of infection, and worms embed themselves in mouse caecum and colon tissue, which closely mimics human whipworm infection and pathology (Klementowicz et al., Reference Klementowicz, Travis and Grencis2012). As with T. trichiura, T. muris remain within the large intestine (which includes the caecum and colon) for their lifespan, and this has resulted in a complex relationship between the parasite and mouse microbiota, where it is known that the parasite manipulates and exploits the gut bacteria to induce persistent infection (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018). It is now apparent that T. muris is dependent on cues from both the host and gut microbiota to initiate infection and generate pathology in the host. Embryonated eggs incubated with gut explants containing caecal bacteria, or bacterial cultures of E. coli (Hayes et al., Reference Hayes, Bancroft, Goldrick, Portsmouth, Roberts and Grencis2010) and/or Staphylococcus aureus (Koyama, Reference Koyama2013) were shown to provide important microbial cues that enable successful worm larvae (stage one, L1) hatching to occur at the start of infection. Hayes et al. (Reference Hayes, Bancroft, Goldrick, Portsmouth, Roberts and Grencis2010) also found that the intestinal bacteria formed aggregates around egg poles, and that this interaction was mediated by adhesion organelles on the surface of bacterial cell membranes, namely type 1 fimbria. Hatching of embryonated eggs from the closely related whipworm T. suis (pig trichuriasis) did not occur using the same bacterial species (both Gram-negative Escherichia coli strains and Gram-positive Lactobacilli, Streptococci and Enterococci strains) that induce T. muris hatching (Vejzagić et al., Reference Vejzagić, Adelfio, Keiser, Kringel, Thamsborg and Kapel2015), suggesting that host–worm–microbiota specific interactions exist. Antibiotic treatment prior to T. muris infection resulted in a significant depletion of gut microbiota that reduced hatching rates, further demonstrating a dependence of Trichuris on the microbiota to initiate infection (Hayes et al., Reference Hayes, Bancroft, Goldrick, Portsmouth, Roberts and Grencis2010).
The interaction of Trichuris muris and the gut microbiota
Many enteric parasites have a tropism for the large intestine, which is the same mucosal niche as the majority of bacteria in the gut microbiota. Within this niche, Trichuris and the gut microbiota are likely to interact with each other through either direct or indirect mechanisms, and it is this interaction and relationship that can influence each other's growth and survival in the mammalian GI tract. Trichuris muris infection protects immunodeficient mice (Nod2−/−) from intestinal abnormalities associated with this IBD-mouse model by enhancing the growth of protective bacterial populations like the Clostridiales, and in turn inhibiting pro-inflammatory species (Ramanan et al., Reference Ramanan, Bowcutt, Lee, Tang, Kurtz, Ding, Honda, Gause, Blaser, Bonneau, Lim, Loke and Cadwell2016). What mechanisms are responsible for inducing changes in the mouse gut microbiota composition (in terms of abundance and diversity) and how they subsequently induce anti-inflammatory responses in the host are largely unknown. We propose mechanisms for altering the gut microbiota may include the physical presence of whipworms in the lumen (worms embedded in intestinal epithelial cells may disrupt surface cell adhesions for bacteria and possible competition for space), secreted worm-derived compounds, competition for nutrients against members of the gut microbiota, and/or worm-induced changes in the host during infection that alter the local GI immune (Th1 or Th2) environment that may also affect the survival of members within the gut microbiota (Fig. 1).
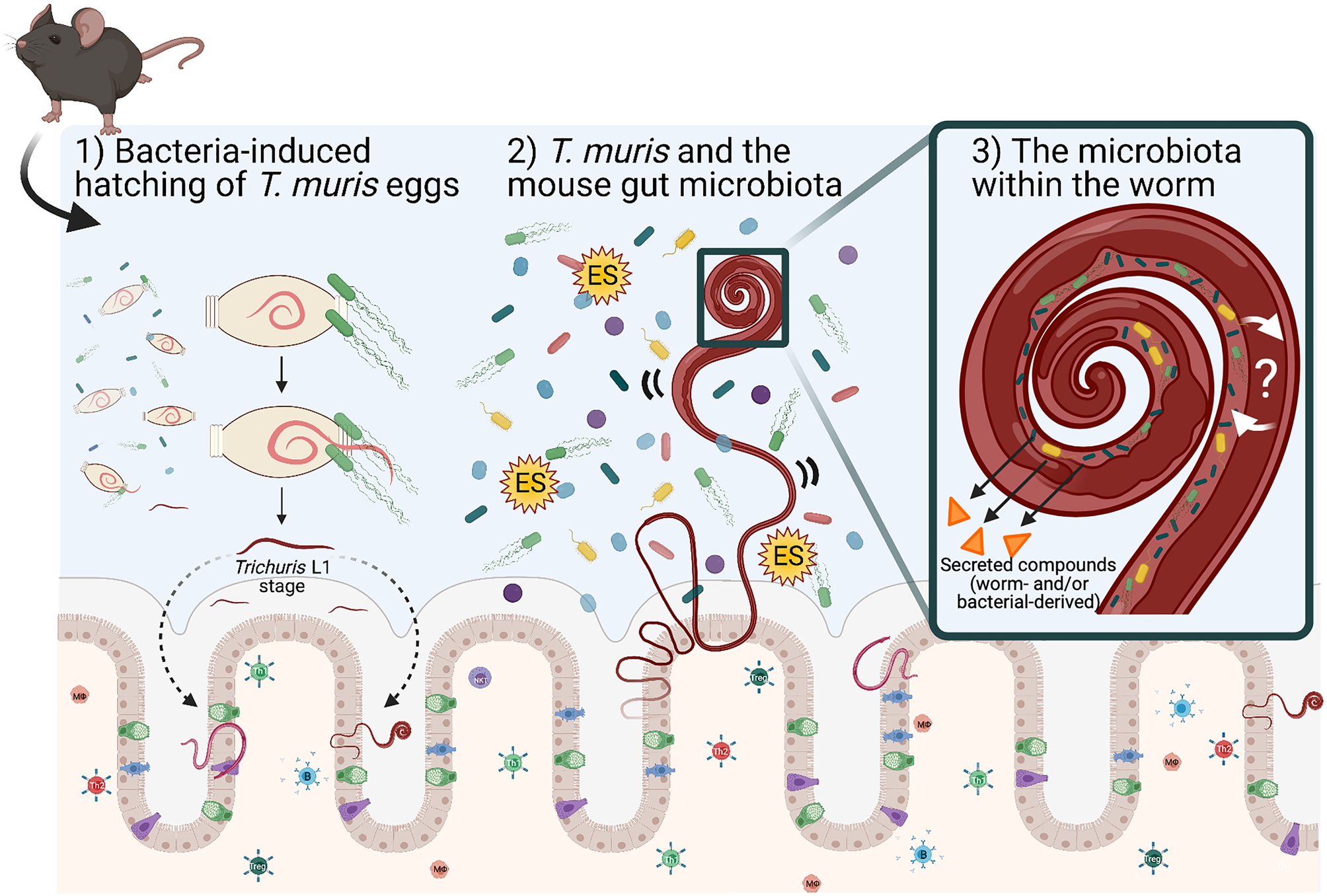
Fig. 1. Schematic representing known and proposed mechanisms of how T. muris interacts with the large intestinal microbiota. (1) Upon entry into the large intestine, worm eggs interact with bacteria to promote hatching and initiate infection of the large intestine. (2) Once embedded in host epithelium, T. muris is proposed to influence the composition of the mouse gut microbiota by direct and indirect mechanisms (competition for space and/or nutrients for growth, by the secretion of metabolites including Trichuris ES and bacterial-derived compounds). (3) T. muris has also evolved to have its own worm gut microbiota, which is derived from the mouse gut microbiota. The identity and function of worm-selective pressures on their gut microbiota, and how the worm-specific bacteria interact with the worm host are unknown. Schematic shows the gut epithelial barrier with epithelial cells (beige), Goblet cells (green), Tuft cells (blue) and enteroendocrine cells (purple) covered in thick mucus (grey). Image created with BioRender.com.
In the absence of all microorganisms, as observed in germ-free mice, a genetically susceptible host becomes resistant to T. muris infection using either embryonated eggs or in vitro hatched L1 larvae (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018). On the other end of the spectrum, laboratory mice rewilded into the environment develop a more diverse microbiota which renders them more susceptible to T. muris infection with greater parasite burdens and worm biomass than their laboratory-dwelling counterparts (Leung et al., Reference Leung, Budischak, Chung The, Hansen, Bowcutt, Neill, Shellman, Loke and Graham2018).
Another study of rewilded mice found that the gut microbiota of T. muris-infected rewilded mice promoted the expansion of Firmicutes, Bacteroidetes, Proteobacteria phyla, and this compositional change was associated with a reduction in colonization resistance against other invading species (specifically members from the Enterobacteriaceae and Rikenellaceace families) when compared to rewilded uninfected mice (Bär et al., Reference Bär, Leung, Hansen, Loke, Hall, Conour and Graham2020). These findings provide further evidence of how important the microbiota is to whipworm infection, and how delicate the balance that exists between microbial diversity (in terms of both composition and abundance) and the severity and persistence for successful worm infection.
Using a well-established model of chronic worm infection, when laboratory wild-type mice are given a single low dose of embryonated eggs (20–30 T. muris eggs) upon transition from larvae to adult worms (day 28 post infection), the gut microbiota undergo compositional changes (Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015; Schachter et al., Reference Schachter, Alvarinho de Oliveira, da Silva, de Barros Alencar, Duarte, da Silva, Ignácio and Lopes-Torres2020). Specifically, bacterial taxa like Firmicutes, Mucispirillum and Proteobacteria increase as a result of infection; whilst Parabacteroides and Prevotella decrease in abundance (Holm et al., Reference Holm, Sorobetea, Kiilerich, Ramayo-Caldas, Estellé, Ma, Madsen, Kristiansen and Svensson-Frej2015; Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015; Schachter et al., Reference Schachter, Alvarinho de Oliveira, da Silva, de Barros Alencar, Duarte, da Silva, Ignácio and Lopes-Torres2020). The observed changes in microbial composition from infection appear to be parasite-dependent and transient in nature. In addition, this altered gut environment makes the host less favourable for re-infection with Trichuris (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018), but whether or not it makes the microbiota more permissive to other infections, like Salmonella (as observed with infection of the small intestinal parasite Heligmosomoides polygyrus bakerii), has yet to be studied (Brosschot et al., Reference Brosschot, Lawrence, Moeller, Kennedy, FitzPatrick, Gauthier, Shin, Gatti, Conway and Reynolds2021). However, three months post infection when no mature whipworms are detectable, the gut microbiota regains bacterial diversity equivalent to their uninfected counterparts (Holm et al., Reference Holm, Sorobetea, Kiilerich, Ramayo-Caldas, Estellé, Ma, Madsen, Kristiansen and Svensson-Frej2015; Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015).
We know that the absence of a microbiota (germ-free mice) prevents Trichuris hatching and infection, whilst the presence of a highly diverse microbiota actually enhances host susceptibility to infection. However, more research is needed to understand how microbial diversity influences worm persistence and survival in the gut, and what role the microbiota may have in manipulating the host immune system response against the invading worms. It is well documented that susceptibility to worm infection is largely dictated by the host immune system response against parasitic infection; CD4+ T helper cell 2 (Th2) responses mount a targeted immune response which leads to rapid worm expulsion, whereas if the host mounts a CD4+ T helper cell 1 response against whipworm infection, the host experiences chronic infections with low worm burdens (Else et al., Reference Else, Hültner and Grencis1992). This is in contrast to other helminth infections where parasite-derived ES products activate tuft cells to initiate Th2 responses, and specifically tuft cell hyperplasia in the gut during chronic infections (Gerbe et al., Reference Gerbe, Sidot, Smyth, Ohmoto, Matsumoto, Dardalhon, Cesses, Garnier, Pouzolles, Brulin, Bruschi, Harcus, Zimmermann, Taylor, Maizels and Jay2016). Germ-free data from enteric roundworm infection using either antibiotic-treated of L3 H. polygyrus larvae or with eggs hatched using an auxotrophic E. coli strain (HA107 that cannot replicate in mice without exogenous supplementation of microbial amino acids) found that worms had impaired fitness as noted by reduced fecundity, and the germ-free hosts had mounted a more robust type 2 humoral responses in the absence of gut bacteria (Rausch et al., Reference Rausch, Midha, Kuhring, Affinass, Radonic, Kühl, Bleich, Renard and Hartmann2018; Russell et al., Reference Russell, Faubert, Verdu and King2020) implicating a possible function of the microbiota in buffering host immune responses to worm infections. Mouse models with varying degrees of microbial diversity (from rewilded to single species colonization of germ-free mice) highlight how critical the composition of the gut microbiota is for Trichuris infection. Manipulation of the gut microbiota in mice to contain genetically altered bacteria may shed light on how these bacteria interact with Trichuris and what functions different genera of bacteria have that contribute to successful whipworm infection.
Reoccurring Trichuriasis and the gut microbiota
A limited number of field studies have been performed investigating the relationship between intestinal helminth infections and the host microbiota; although there are conflicting reports. A field study examining the microbiota composition of Ecuadorian children found T. trichuria infection did not induce significant changes in the gut microbiota composition (Cooper et al., Reference Cooper, Walker, Reyes, Chico, Salter, Vaca and Parkhill2013), whereas individuals infected with intestinal helminths (including Trichuris) within the Malaysian indigenous (Orang Asli) populations were found to have higher microbial diversity than their negative counterparts (Lee et al., Reference Lee, Tang, Lim, Choy, Kurtz, Cox, Gundra, Cho, Bonneau, Blaser, Chua and Loke2014). It is possible that the conflicting data in human field studies, and their comparison to mouse models are due to the fact that infected individuals are rarely exposed to a single, low-dose helminth infection but rather continuous re-occurring infection by the same or multiple parasites. A recent study in Tanzania, was one of the first to identify a reduction in the abundance of gut bacteria: Bacteroidetes, Proteobacteria and Actinobacteria associated with a cohort of women and children solely infected with T. trichuria (Chen et al., Reference Chen, Mozzicafreddo, Pierella, Carletti, Piersanti, Ali, Ame, Wang and Miceli2021); which is similar to the microbial changes reported in mouse studies (Holm et al., Reference Holm, Sorobetea, Kiilerich, Ramayo-Caldas, Estellé, Ma, Madsen, Kristiansen and Svensson-Frej2015; Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015). Parallels between human and mouse studies were further addressed by identifying bacterial taxa that increased in both T. trichuria-only infected Indonesian individuals and T. muris-infected mice, these genera included Bacteroides, Collinsella, Subdoligranulum, Escherichia/Shigella, Prevetoella and Streptococcus (Rosa et al., Reference Rosa, Snowden, Martin, Fischer, Kupritz, Beshah, Supali, Gankpala, Fischer, Urban and Mitreva2021). There has also been the development of a mouse model to partially address the discrepancy in human and mouse data that involves a trickle model of T. muris infection, where mice are routinely (weekly) infected with low doses of embryonated eggs, thereby closely mimicking the low-dose chronic human infection by T. trichiura (Glover et al., Reference Glover, Colombo, Thornton and Grencis2019). This simulated model of natural infection with repeated exposure to T. muris caused a decrease in microbial diversity early on during infection that rapidly resolved over time to control levels upon repeated exposure (Glover et al., Reference Glover, Colombo, Thornton and Grencis2019). Interestingly, this tickle model was associated with the development of partial protective immunity supporting the concept of a complex interplay between host immunity, parasite and microbiota. Overall, laboratory experiments and field studies indicate that primary infection by Trichuris influences the microbiota composition, but the true nature of the relationship between the microbiota and intestinal parasites remains largely unknown.
Trichuris muris’ intimate relationship with bacteria
While multiple studies suggest that the large metazoan parasite Trichuris has evolved to respond to microbial cues in the gut to initiate hatching (and infection) of the host, mature whipworms have also been shown to harbour their own unique gut microbiota by 16S rRNA gene denaturing gel electrophoresis and fluorescence in situ hybridization (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018). Irrespective of the host microbiota composition, mature whipworms acquire a distinct microbiota selected from members residing in the host gut during infection (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018). The worm microbiota is largely dominated by bacterial species belonging to the phylum Proteobacteria (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018), which begs the questions as to how does the worm select these bacteria, why Proteobacteria and what role do these particular bacteria provide for worms? Currently, worm-specific selective pressures on bacteria are unknown, and whether the bacteria are selected based on their function in the gut microbiota, their ability to adapt to new environments (such as the worm gut) or if they are exploited as an energy source (food) still need to be answered. The fact whipworms evolve and generate their own microbiota adds complexity to the host–parasite–microbiota relationship and raises new questions in understanding each of the mutualistic relationships present in the gut. Future work may include using a defined model system of a single microbial species in a germ-free host with the nematode Trichuris to address these outstanding questions, simply because of the many advantages this system has to offer in terms of experimental tractability for exploring the relationship between Trichuris and the microbiota.
Metabolites as moieties for cross-talk
Trichuris has developed effective strategies against the host immune system by exploiting its immunoregulatory properties to permit chronic infection; identification of potential candidates in this cross-talk is likely to be metabolites produced by (directly or indirectly) the worm during infection. In chronic infection, T. muris worms reach maturity at day 33 post infection, at which point mucosal immune and pathological changes in response to infection have occurred, and the microbial population and the worm have adapted to each other. Faecal metabolite analysis at day 41 of chronic infection identified increased concentrations of amino acids including phenylalanine, serine, threonine, leucine, ornithine and glycine, with a decrease in the abundance of vitamin D2/D3 derivatives, fatty acid and glycerophospholipid metabolites, compounds involved in heme degradation and metabolites involved in the biosynthesis of tryptophan, lysine, cystine and arginine amino acids when compared to naïve samples (Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015). This altered metabolic profile can be interpreted as a hallmark of the skewed gut microbiota due to chronic infection and the ability of the microbiota to differentially harvest resources from the host diet. What is currently unclear is the origin of these observed metabolite changes and the relative contributions of the host, parasite and microbiota.
A reduction in luminal SCFA butyrate occurs during infection, and in turn host cells downregulate the expression of butyrate transporters, suggesting a transition to utilising other readily available energy sources available in the gut (White, Reference White2016). Bacteria are the major source of SCFA in the gut, specifically the abundance and function of butyrate producers including taxa within the Firmicutes phylum like Clostridiales clusters IV and XIVa, and Ruminococcaceae (Louis et al., Reference Louis, Young, Holtrop and Flint2010), as well as members of Bacteroidetes, Actinobacteria and Proteobacteria that produce and utilize butyrate for the synthesis of essential amino acids are able to influence host health. Many of these bacteria are reduced in number during worm infection, and contribute to the alterations of microbial-derived metabolites like butyrate (Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015). Trichuris is unable to synthesize butyrate, and supplementation of butyrate during chronic infection had minimal impact on worm burden or disease severity (Colombo and Grencis, unpublished data). Interpretation of this data may suggest that the worm presence and infection in the large intestine induces a mucosal inflammatory response that makes the gut less favourable for the growth of butyrate-producing bacteria like Clostridia and Ruminococcacae that accounts for the reduction in butyrate-regulated FoxP3+ CD4+ Treg cells in the colonic lamina propria (Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015).
Trichuris-specific metabolites
Trichuris muris eggs are devoid of bacteria (White et al., Reference White, Houlden, Bancroft, Hayes, Goldrick, Grencis and Roberts2018); therefore, at this life cycle stage, all identified metabolites are likely to be worm origin (rather than microbial). Using mass spectrometry, T. muris-egg specific metabolites were identified to belong to amino acid metabolism, carbohydrate metabolism and nucleotide metabolism (Yeshi et al., Reference Yeshi, Creek, Anderson, Ritmejerytė, Becker, Loukas and Wangchuk2020). Within 90 min of entering the gut, the first-stage larvae are released from the egg and begin to borough into the epithelium; at which point the worms begin to grow and mature and release proteins into the niche called excretory–secretory (ES) products (Bancroft et al., Reference Bancroft, Levy, Jowitt, Hayes, Thompson, Mckenzie, Ball, Dubaissi, France, Bellina, Sharpe, Mironov, Brown, Cook, S MacDonald, Thornton and Grencis2019). In contrast to other parasitic metabolites, such as those produced by Nippostrongylus brasiliensis infection; Trichuris ES-enriched fractions contain an abundance of fatty acids including SCFA, amino acids and enriched organic compounds (Wangchuk et al., Reference Wangchuk, Kouremenos, Eichenberger, Pearson, Susianto, Wishart, McConville and Loukas2019). Specifically, lactic acid, oleic acid, succinate acid, the SCFAs: propionate, butyrate and acetate; alanine, glutamine, sugars: fructose, galactose, glucosamine and glucose-6-phosphate; and adenine and uridine (nucleobases/nucleosides) were found to be increased (Wangchuk et al., Reference Wangchuk, Kouremenos, Eichenberger, Pearson, Susianto, Wishart, McConville and Loukas2019). However, T. muris lacks known gene homologs for the synthesis of these metabolites, for example, there are no gene clusters identified to generate de novo synthesis of SCFA butyrate in the worm genome (White, Reference White2016). Therefore, the identified metabolites by Wangchuk et al. (Reference Wangchuk, Kouremenos, Eichenberger, Pearson, Susianto, Wishart, McConville and Loukas2019) are more likely to be a combination of Trichuris-derived metabolites and metabolic compounds generated by their own gut microbiota, which adult worms acquire from the host microbiota.
In chronic infection, the immune system generates a ‘tolerogenic-like’ immune response against the worms, which is likely to be mediated from a number of Trichuris-specific anti-inflammatory metabolites increased in ES-fractions including glucosamine, uridine and butyrate (Wangchuk et al., Reference Wangchuk, Kouremenos, Eichenberger, Pearson, Susianto, Wishart, McConville and Loukas2019). High levels of succinate and lactic acid were found in the ES-fraction, and the immunoregulatory effect of butyrate, succinate and lactic acid may have a role in contributing to anaerobic metabolism in the caecum, which would reflect a possible beneficial trait of the worm gut microbiota (Wangchuk et al., Reference Wangchuk, Kouremenos, Eichenberger, Pearson, Susianto, Wishart, McConville and Loukas2019). Extracellular vesicles (EV) also present in the ES faction of T. muris are internalized by the host cells and been found to contain a multitude of proteins including those involved in proteolysis, mRNAs and microRNAs (Eichenberger et al., Reference Eichenberger, Talukder, Field, Wangchuk, Giacomin, Loukas and Sotillo2018). Recently, using mouse caecaloids (organoids made from the caecum), it has been shown that T. muris-specific EVs downregulate responses to nucleic acid recognition and type 1 interferon immune-mediated responses (Duque-Corre et al., Reference Duque-Correa, Schreiber, Rodgers, Goulding, Forrest, White, Forrest, White, Buck, Grencis and Berriman2020). How worm-specific EVs interact with the microbiota is unclear (Eichenberger et al., Reference Eichenberger, Talukder, Field, Wangchuk, Giacomin, Loukas and Sotillo2018). Not only does the identification of T. muris-secreted metabolites during infection provide a tool for understanding how whipworms interact with both the host and bacterial environment, but also may lead to the design of novel approaches to controlling chronic worm infections.
Treatments
A variety of anthelmintic drugs are used to treat Trichuris infections in humans and livestock. For T. muris infection anthelmintic drugs include, but are not limited to the administration of albendazole, mebendazole, levamisole, pyrantel pamoate and ivermectin that are also commonly used to treat T. trichiura infections (Keiser et al., Reference Keiser, Tritten, Adelfio and Vargas2012). How these drugs impact the gut microbiota in the host and worm remains largely elusive. Houlden et al. (Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015) found mebendazole treatment on naïve mice did not influence the composition of the gut microbiota, however many of the other drugs have yet to be studied. A reduction in microbial species diversity was observed in antibiotic-treated mice prior to infection that led to both reduced worm burden in the mice, and a modulated T-cell-dependent immune response mounted against existing worms (Hayes et al., Reference Hayes, Bancroft, Goldrick, Portsmouth, Roberts and Grencis2010; Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015). Transient antibiotic treatment of mice prior to a high-dose T. muris infection led to enhanced susceptibility to infection (Scott et al., Reference Scott, Andrusaite, Andersen, Lawson, Alcon-Giner, Leclaire, Caim, Le Gall, Shaw, Connolly, Roe, Wessel, Bravo-Blas, Thomson, Kästele, Wang, Peterson, Bancroft, Li, Grencis, Mowat, Hall, Travis, Milling and Mann2018) and it was proposed that this was dependent upon the generation of dysregulated macrophages promoting type 1 immune responses, again supporting a complex interplay between host and microbiota influencing infection outcome. As an alternative to drug therapy, repeated administration of live beneficial bacteria like the probiotic strain Lactobacillus rhamnosus (isolate JB-1) caused an acceleration of worm expulsion in mice, mediated by an IL-10/Goblet cell pathway induced in the murine host (McClemens et al., Reference McClemens, Kim, Wang, Mao, Collins, Kunze, Bienenstock, Forsythe and Khan2013). Manipulation of the host microbiota with probiotics to reduce worm burden further highlights the intricate relationship between the gut microbiota and whipworms during infection.
In humans, there is limited and conflicting information regarding the impact of anthelmintic treatment on the gut microbiota. The majority of infected individuals have worm burdens below clinical thresholds, and that can remain in their host for years contributing to poor nutrition and impaired growth and development, particularly in children (Callender et al., Reference Callender, Grantham-Mcgregor, Walker and Cooper1992; Nokes et al., Reference Nokes, Grantham-McGregor, Sawyer, Cooper and Bundy1992). It is unclear if the enhanced burden of chronic whipworms or the altered gut microbiota associated with these infections is the cause or effect of these health concerns in individuals, and if employing long-term administration of prebiotic and/or probiotic treatments to alter the gut microbiota would be a step forward in controlling parasitic infections.
Conclusions
The importance of the microbiota during helminth infection is becoming increasingly recognized, and is also the start of an exciting and crucial area of parasitological research. Microbial changes have been reported for multiple intestinal helminths including infections with Ascaris lumbricoides (Ramírez-Carrillo et al., Reference Ramírez-Carrillo, Gaona, Nieto, Sánchez-Quinto, Cerqueda-García, Falcón, Rojas-Ramos and González-Santoyo2020), H. polygyrus (Walk et al., Reference Walk, Blum, Ewing, Weinstock and Young2010; Kreisinger et al., Reference Kreisinger, Bastien, Hauffe, Marchesi and Perkins2015), Necator americanus (hookworm; Cantacessi et al., Reference Cantacessi, Giacomin, Croese, Zakrzewski, Sotillo, McCann, Nolan, Mitreva, Krause and Loukas2014) and of interest for this review T. trichiura (Lee et al., Reference Lee, Tang, Lim, Choy, Kurtz, Cox, Gundra, Cho, Bonneau, Blaser, Chua and Loke2014; Chen et al., Reference Chen, Mozzicafreddo, Pierella, Carletti, Piersanti, Ali, Ame, Wang and Miceli2021) and T. muris (Holm et al., Reference Holm, Sorobetea, Kiilerich, Ramayo-Caldas, Estellé, Ma, Madsen, Kristiansen and Svensson-Frej2015; Houlden et al., Reference Houlden, Hayes, Bancroft, Worthington, Wang, Grencis and Roberts2015; Schachter et al., Reference Schachter, Alvarinho de Oliveira, da Silva, de Barros Alencar, Duarte, da Silva, Ignácio and Lopes-Torres2020).
Using novel tools such as germ-free models to decipher the intricate relationship that the microbiota has with whipworms opens new perspectives on how these helminths function and infect their hosts. In addition, field studies of re-wilding mice, or potentially wild, naturally occurring infection mouse models provide evidence into the complexity and dependence of the gut microbiota and T. muris, and possible insight into T. trichuria infections. Helminth metabolites are also likely to be candidates for regulating the immune system, positively contributing to microbial richness and diversity in individuals who suffer from chronic inflammatory diseases, or in turn as potential candidates for anthelminthic drugs (as discussed in Garcia-Bustos et al., Reference Garcia-Bustos, Sleebs and Gasser2019 for Caenorhabditis elegans). Harnessing how enteric parasites, such as whipworms, interact and modulate the gut microbiota will open new avenues for exploring anthelminthic treatments and addressing the growing concern of the ‘microbial diversity’ hypothesis regarding the dramatic increase in the incidence of allergic and autoimmune diseases in developed countries. Human trials using Trichuris eggs (specifically T. suis) show promise as a probiotic agent, and has been used in trials as a treatment for multiple sclerosis (Fleming et al., Reference Fleming, Isaak, Lee, Luzzio, Carrithers, Cook, Field, Boland and Fabry2011, Reference Fleming, Hernandez, Hartman, Maksimovic, Nace, Lawler, Risa, Cook, Agni, Reichelderfer, Luzzio, Rolak, Field and Fabry2019; Yordanova et al., Reference Yordanova, Ebner, Schulz, Steinfelder, Rosche, Bolze, Paul, Mei and Hartmann2021; Williams et al., Reference Williams, Dige, Rasmussen, Hvas, Dahlerup, Iversen, Stensvold, Agnholt and Nejsum2017). More recently, clinical intervention trials using T. suis eggs against psoriasis and IBD are ongoing in North America, and it will be interesting to see how Trichuris can reduce symptoms mediated by an altered immune response and potentially by changes in gut microbial composition (Prosberg and Petersen, Reference Prosberg and Petersen2018–2021). Understanding how Trichuris interacts with the gut microbiota during infection is of particular interest, as it is the organ that harbours the largest and most diverse microbial community within our bodies. In conclusion, we anticipate there will be substantial growth in this field and more research examining how to harness this information to develop new helminth therapies that control the spread and infection in humans.
Author contributions
M.A.E.L. conducted the literature search, designed and wrote the review and figure. R.K.G. and I.S.R. provided feedback and comments on the manuscript.
Financial support
M.A.E.L. and R.K.G. are funded by Wellcome Trust investigator award (Z10661/Z/18/Z awarded to RKG). The Wellcome Trust Centre for Cell Matrix Research, University of Manchester is also supported by centre funding from the Wellcome Trust (203128/Z/16/Z).
Conflict of interest
None.
Ethical standards
Not applicable.