INTRODUCTION
Plasmodium knowlesi, a parasite adapted to Southeast Asian macaques, occupies a unique position among the Plasmodium species. Not only is P. knowlesi pathogenic when it crosses from the natural macaque hosts into the human population, it is also permissive in a range of experimental non-human primates (NHPs). In deed P. knowlesi is a parasite well known to the malaria research community and has been used extensively as a model for parasite biology, drug and vaccine development since 1932, reviewed in this issue (Butcher and Mitchell, Reference Butcher and Mitchell2016; Pasini et al. Reference Pasini, Zeeman, Voorberg-VAN DER Wel and Kocken2016; Galinski et al. Reference Galinski, Lapp, Peterson, Ay, Joyner, LE Roch, Fonseca, Voit and Mahpic2017). Juxtaposed to research, P. knowlesi was virtually unknown to contemporary medical practitioners until 2004 when a large focus of naturally acquired P. knowlesi malaria was reported in the human population of Sarawak, Malaysian Borneo (Singh et al. Reference Singh, Kim Sung, Matusop, Radhakrishnan, Shamsul, Cox-Singh, Thomas and Conway2004). Remarkably detailed information on several experimental lines of P. knowlesi existed long before P. knowlesi was recognized as an emerging pathogen in the human population of Southeast Asia.
To put the new millennium find into context, the last of the four Plasmodium species associated with malaria in the human population, Plasmodium ovale, was described in 1922 (Coatney et al. Reference Coatney, Collins, Warren, Contacos and Coatney1971a ). The other human-host adapted species are Plasmodium falciparum, Plasmodium vivax and Plasmodium malariae. Of these, P. falciparum is the most virulent and prevalent species responsible for most of the estimated 212 million (range 148–304 million) new cases of malaria and 429 000 (range 235 000–639 000) malaria-related deaths, in 2015 (WHO, 2016).
Consequently, malaria research was, and still is, underpinned by the quest to understand, control and prevent malaria caused by P. falciparum. In addition to virulence as a distinguishing feature, P. falciparum has other unique features when compared to the other human-host adapted Plasmodium species, the most striking is binding of P. falciparum-infected red blood cells to host endothelium, recently reviewed (Mosnier and Lavstsen, Reference Mosnier and Lavstsen2016; Wassmer and Grau, Reference Wassmer and Grau2017). While some of the unique characters are more compelling virulence candidates than others, all are difficult to test.
EXPERIMENTAL MODEL AND ZOONOTIC PATHOGEN
Providing experimentally sound evidence that particular characteristics of P. falciparum are indeed related to virulence and taking these forward to develop treatments and augmentative therapies require representative model systems, including modelling in vivo. Indeed, an in vivo model system remains the missing link in translational research on malaria pathophysiology. Even though the falciparum clade of Plasmodium species has recently had new members added, including the multiple variants of Plasmodium praefalciparum, Plasmodium billcollinsi, Plasmodium blacklocki, Plasmodium adleri and Plasmodium gaboni, P. falciparum isolated from human infections is more or less resolutely human host restricted (Loy et al. Reference Loy, Liu, Li, Learn, Plenderleith, Sundararaman, Sharp and Hahn2017). On this back-drop and, despite huge research efforts to understand the pathophysiology of severe malaria, outputs remain largely hypothetical.
The very recent finding that, like P. falciparum, P. knowlesi causes severe and fatal infections in the human population opens the possibility for comparative research on malaria parasite virulence. Importantly, and in contrast to P. falciparum, P. knowlesi is experimentally permissive in a range of NHPs, conferring a means to test virulence (Cox-Singh and Culleton, Reference Cox-Singh and Culleton2015). Is it possible that P. knowlesi may help resolve what is effectively a research bottleneck?
THE PROBLEM WITH MODELS
Phylogenetically P. knowlesi falls within the old-world monkey clade, a clade that includes P. vivax, and is evolutionarily relatively distant to P. falciparum. Perceived wisdom built on Plasmodium phylogenies determined that P. knowlesi, while a valuable model for parasite biology, was considered too evolutionarily distant from P. falciparum to model for malaria pathophysiology and considered more suitable to model for P. vivax (Craig et al. Reference Craig, Grau, Janse, Kazura, Milner, Barnwell, Turner and Langhorne2012). A view supported by Martinelli and Culleton, this issue (Martinelli and Culleton, Reference Martinelli and Culleton2016).
In addition, the most common experimental partner for P. knowlesi is the Rhesus macaque and, while used extensively, this model is not optimal to represent severe malaria in the human host. This is because most contemporary laboratory colonies of Rhesus macaques do not mount functional resistance to experimental infections with P. knowlesi (Praba-Egge et al. Reference Praba-Egge, Montenegro, Cogswell, Hopper and James2002). Consequently Rhesus monkeys infected with P. knowlesi develop an overwhelming parasitaemia and a disease phenotype that is difficult to reconcile with severe P. falciparum or even, latterly, severe P. knowlesi infections in the human host (Cox-Singh and Culleton, Reference Cox-Singh and Culleton2015).
While the hyper-susceptible phenotype in Rhesus macaques re-enforced the perception that P. knowlesi is not a suitable model for malaria pathophysiology this phenotype is not universal among Rhesus macaques, as Butcher and Mitchell point out in this issue (Butcher and Mitchell, Reference Butcher and Mitchell2016). Originally there were several P. knowlesi – Rhesus macaque models and early research described variable disease phenotypes depending on the experimental P. knowlesi line used and the origin of the Rhesus monkey (Butcher and Mitchell, Reference Butcher and Mitchell2016). With time, the highly susceptible Rhesus of Indian origin became the experimental host of choice, and information on diverse disease phenotypes was effectively lost. Furthermore, not all of the earlier laboratory adapted lines of P. knowlesi survived. The Indian macaque partnered with a few experimental lines of P. knowlesi was established and resulted in an homogenized model system that did not represent severe falciparum malaria. Taken together with the positioning of P. knowlesi relatively close to P. vivax in the Plasmodium phylogenies, the notion that P. knowlesi could not model for severe falciparum malaria became ingrained.
THE PROBLEM WITH DOGMAS
Perceived wisdom can be difficult to change. Even reports of a severe disease phenotype in Olive baboons, experimentally infected with P. knowlesi, with compelling pathological features similar to severe P. falciparum malaria with coma and latterly placental malaria in the same model, did not sway the accepted opinion that P. knowlesi is a more representative model for P. vivax (Ozwara et al. Reference Ozwara, Langermans, Maamun, Farah, Yole, Mwenda, Weiler and Thomas2003; Onditi et al. Reference Onditi, Nyamongo, Omwandho, Maina, Maloba, Farah, King, Moore and Ozwara2015).
By extension of the same logic, leaning on phylogeny, P. knowlesi biology is most often compared with P. vivax, even though P. knowlesi and P. vivax bear little morphological or biological resemblance to each other. For example, hypnozoite liver stages, reticulocyte restriction and enlargement of the infected red blood cell, characteristic of P. vivax are absent in P. knowlesi as are Schuffner's stippling (Coatney et al. Reference Coatney, Collins, Warren, Contacos and Coatney1971b ). We now know that P. knowlesi can cause severe acute malaria in the human population similar to P. falciparum and furthermore both species express antigenically variant surface antigens on the infected host red blood cell (Brown and Brown, Reference Brown and Brown1965; Howard et al. Reference Howard, Barnwell and Kao1983). This type of antigenic variation is the product of complex multiple gene families present in both P. falciparum, the var gene family, and P. knowlesi, the SICAvar gene family, and absent in P. vivax (Baruch et al. Reference Baruch, Pasloske, Singh, Bi, Ma, Feldman, Taraschi and Howard1995; Smith et al. Reference Smith, Chitnis, Craig, Roberts, Hudson-Taylor, Peterson, Pinches, Newbold and Miller1995; Su et al. Reference Su, Heatwole, Wertheimer, Guinet, Herrfeldt, Peterson, Ravetch and Wellems1995; al-Khedery et al. Reference al-Khedery, Barnwell and Galinski1999). In P. falciparum, the var gene family is strongly implicated in virulence (Mosnier and Lavstsen, Reference Mosnier and Lavstsen2016; Wassmer and Grau, Reference Wassmer and Grau2017). Although in P. knowlesi the role of the likely orthologous SICAvar gene family in precipitating human disease is not yet known, the fact that both species cause severe disease and both have this gene family deserves further investigation. Even more compelling, the WHO criteria for severe malaria fit malaria caused by P. falciparum and P. knowlesi. The important exception is that, so far, patients with severe malaria caused by P. knowlesi do not deteriorate into the classical severe malaria with coma disease phenotype, even though parasite-infected erythrocytes were observed in the brain microvasculature of a fatal case of P. knowlesi malaria (Fig. 1) (Cox-Singh et al. Reference Cox-Singh, Hiu, Lucas, Divis, Zulkarnaen, Chandran, Wong, Adem, Zaki, Singh and Krishna2010; Barber et al. Reference Barber, Rajahram, Grigg, William and Anstey2017; Daneshvar et al. Reference Daneshvar, William and Davis2017). Nonetheless, positioning in genetic phylogeny appears to carry more weight than clinical and biological evidence. Perhaps we need to re-consider how we identify and rank the characteristics of representative models for malaria pathophysiology.
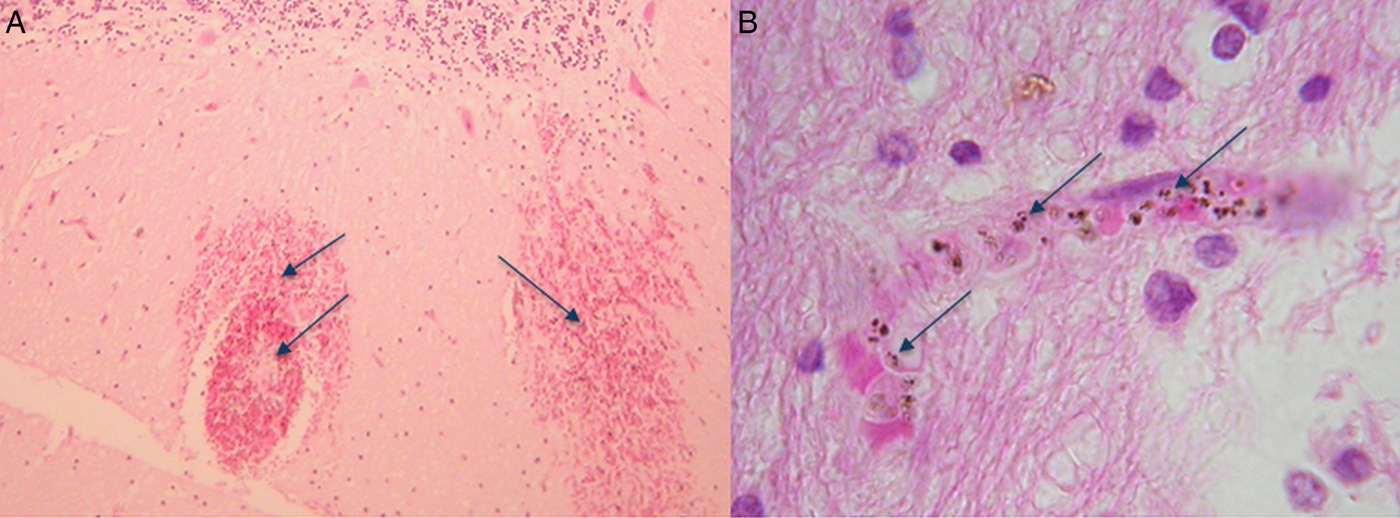
Fig. 1. Haematoxylin and eosin-stained sections from the brain of a fatal case of Plasmodium knowlesi malaria. (A) Haemorrhages in white matter containing pigmented parasites (arrows) and (B) at higher magnification showing a capillary congested with pigmented parasite-infected erythrocytes (arrows). The patient was not in coma characteristic of severe falciparum malaria (for detailed descriptions, see Cox-Singh et al. Reference Cox-Singh, Hiu, Lucas, Divis, Zulkarnaen, Chandran, Wong, Adem, Zaki, Singh and Krishna2010).
CHANGING LANDSCAPES AND MOVING ON
Perhaps it is time to consider P. knowlesi as a candidate to model for malaria pathophysiology for the following reasons: P. knowlesi is an experimental model parasite in vivo that is permissive in a number of NHP hosts (Pasini et al. Reference Pasini, Zeeman, Voorberg-VAN DER Wel and Kocken2016); P. knowlesi has been recently adapted to in vitro culture in human erythrocytes (Moon et al. Reference Moon, Hall, Rangkuti, Ho, Almond, Mitchell, Pain, Holder and Blackman2013); P. knowlesi is highly amenable to genetic manipulation (Kocken et al. Reference Kocken, Ozwara, van der Wel, Beetsma, Mwenda and Thomas2002; Moon et al. Reference Moon, Hall, Rangkuti, Ho, Almond, Mitchell, Pain, Holder and Blackman2013); clinical and laboratory data from patients with naturally acquired P. knowlesi malaria are accumulating, and matched information on genetic and genomic diversity to identify and interrogate parasite virulence in vitro and in vivo is available (Daneshvar et al. Reference Daneshvar, Davis, Cox-Singh, Rafa'ee, Zakaria, Divis and Singh2009; Barber et al. Reference Barber, William, Jikal, Jilip, Dhararaj, Menon, Yeo and Anstey2011, Reference Barber, William, Grigg, Menon, Auburn, Marfurt, Anstey and Yeo2013, Reference Barber, Grigg, William, Yeo and Anstey2016; Cox-Singh et al. Reference Cox-Singh, Singh, Daneshvar, Planche, Parker-Williams and Krishna2011; Ahmed et al. Reference Ahmed, Pinheiro, Divis, Siner, Zainudin, Wong, Lu, Singh-Khaira, Millar, Lynch, Willmann, Singh, Krishna and Cox-Singh2014; Pinheiro et al. Reference Pinheiro, Ahmed, Millar, Sanderson, Otto, Lu, Krishna, Rayner and Cox-Singh2015; Lapp et al. Reference Lapp, Geraldo, Chien, Ay, Pakala, Batugedara, Humphrey, Ma, De, Le Roch, Galinski and Kissinger2017). Taken together surely it is time to characterize P. knowelsi, including genetically modified parasites, in several NHP hosts to compare with the differing pathologies that are collectively referred to as severe malaria.
All of that being said, is there an appetite among funders or the malaria research community to support the, by no means trivial, development of a translational in vivo model for malaria pathophysiology? Arguments against this initiative include giving precedence to malaria control rather than defining disease process, an argument further reinforced by the sustainable development goals, Transforming Our World, set by the United Nations in 2016 (https://sustainabledevelopment.un.org/post2015/transformingourworld), goals that aim to end the epidemics of infectious disease and neglected tropical diseases, including malaria, by 2030. A target similar to but less specific than that set in 2000 in the millennium development goals with malaria elimination targets set for 2015 (WHO, 2016). Experience suggests that these targets are difficult to achieve and in the interim, while the number of deaths due to malaria have decreased, just under half of the world population remain at risk of malaria (WHO, 2016). Of course for most that risk is negligible, but for some the risk is very real. Particularly at risk are the world's poorest, especially those under the age of 5 years. Perhaps this argument is enough to support research on malaria pathophysiology, including support to develop an in vivo translational model.
EMERGING INFECTIONS
A word of caution seems prudent – infectious diseases, including newly emergent infections, tend to be of zoonotic origin. The entry of P. knowlesi malaria into the human population in Southeast Asia is a good contemporary example. The entry is on-going and across Southeast Asia. There is no evidence to suggest that malaria caused by P. knowlesi is abating and, in terms of research, we remain several steps behind nature. Vythilingam et al. this issue highlight just how little is known of the vectors and transmission of P. knowlesi (Vythilingam et al. Reference Vythilingam, Wong and Wan-Yussof2016). Yakob et al., also this issue, demonstrate the importance of data on vectors to inform mathematical models to describe P. knowlesi epidemiology (Yakob et al. Reference Yakob, Lloyd, Kao, Ferguson, Brock, Drakeley and Bonsall2017) Contextually, how feasible is the goal to end epidemics if they are zoonotic in origin? And yet ambitious goals are set? A case to include zoonotic malaria in the WHO malaria reports to facilitate policy and malaria control strategies in Southeast Asia was made by Barber et al. (Reference Barber, Rajahram, Grigg, William and Anstey2017). The need to make a case strongly suggests that zoonoses have perhaps, alarmingly, slipped under the policy makers’ radar.
On that backdrop, a dichotomy emerges – funders are willing to support clinical research on severe malaria and research on rodent models for severe disease; yet the same funders are reluctant to engage in discussions on the development of a NHP translational model for severe malaria. Clinical studies collect important data, not least on parasite and host genetic markers of severe disease, markers of immune pathology and immune protection, markers of virulence, yet this work, while valuable, is bottlenecked by the lack of a means to (a) confirm the findings in vivo, (b) identify the triggers and pathways to severe disease and (c) translate the results into human health benefit through the rational development of augmentative therapies designed to protect the severely ill through the acute phase of their illness. Translating research on pathophysiology in any disease requires a representative in vivo model system. At the risk of repetition, a representative in vivo model for severe malaria is not available to perform proof of concept, cause and effective translational research. In the absence of this essential tool, the outputs of clinical and all other research on severe malaria remain and will remain academic. In terms of benefit to communities at risk, money that funds research that cannot be translated could be more beneficial if invested locally to improve access to healthcare, education, safe drinking water and vector control to name but a few.
A POINT OF VIEW
Continued investment in research on severe malaria, which does not include investment in in vivo model development for translational research, is fundamentally flawed. Surely the sole reason to study malaria is to reduce the health impact on communities at risk. If funding and support to develop a vital tool, an in vivo model system, is not forthcoming, we need to question support for on-going research on severe malaria. The outputs of current research that are well placed to inform translational research may not be taken forward. If lack of interest and support to develop a translational model persists, then we run the risk of preserving the academic success of the few while failing the many that include the world's poorest who live with the real threat of developing severe life-threatening malaria. Even more frustrating we have the omics revolution at our disposal and a real opportunity to study and interrogate the host–pathogen interface in exquisite detail. Yet without the means to convert the effort into impact-driven research, progress remains grid-locked.
AN OPPORTUNITY
Articles in this special issue highlight the contribution of research on P. knowlesi to our current understanding of antigenic variation, parasite invasion of host red blood cells, immunity, drug and vaccine development in malaria (Butcher and Mitchell, Reference Butcher and Mitchell2016; Pasini et al. Reference Pasini, Zeeman, Voorberg-VAN DER Wel and Kocken2016; Galinski et al. Reference Galinski, Lapp, Peterson, Ay, Joyner, LE Roch, Fonseca, Voit and Mahpic2017). Undoubtedly the value of P. knowlesi as a laboratory model has waxed and waned over time depending on the needs and particular interests of the research community at any one time. The contemporary finding that P. knowlesi has entered the human population from the natural old-world macaque hosts and causes a severe form of malaria with similarity to P. falciparum would be expected to heighten interest and application of P. knowlesi – the experimental model – to interrogate the many unanswered questions on malaria pathophysiology (Mosnier and Lavstsen, Reference Mosnier and Lavstsen2016; Wassmer and Grau, Reference Wassmer and Grau2017). There are obstacles, the number of contemporary research groups with local access to primate research facilities is very small. Plasmodium knowlesi while, an interesting parasite, is relatively inaccessible, expensive and ethically more challenging to work with than P. falciparum in vitro or the in vivo rodent models for malaria. Despite the obstacles, the opportunities to exploit omics, genetic manipulation, in vitro and in vivo experiments that are informed by human disease are huge. Plasmodium knowlesi must surely be considered a rational choice for a focused, international initiative to develop, characterize and make available a clinically relevant model system, including a well-matched NHP host, for cause and effect studies on severe malaria (Cox-Singh and Culleton, Reference Cox-Singh and Culleton2015; Pasini et al. Reference Pasini, Zeeman, Voorberg-VAN DER Wel and Kocken2016; Galinski et al. Reference Galinski, Lapp, Peterson, Ay, Joyner, LE Roch, Fonseca, Voit and Mahpic2017). We have a wealth of information and hypotheses generated from pain-staking clinical and experimental studies on severe malaria pathophysiology and virulence. Now we need the means to test and translate outputs into health benefit – can P. knowlesi, a pioneer parasite in malaria research, help us conquer this difficult frontier?
ACKNOWLEDGEMENTS
In admiration of all those who have worked on P. knowlesi as a research tool and to those working in the field informing the research community and policy makers on zoonotic malaria. The author is grateful to the Sasakawa Foundation for travel support to initiate collaborative discussions on animal model development in Japan.
DISCLAIMER
Any views expressed in this editorial are the personal views of the author and are not endorsed by any other individual or institution.
COVER
Zoonotic malaria transmission – humans are accidental hosts (Macaca fascicularis image kindly donated by Cyrus Daneshvar); chromosomal synteny adapted from Figure 2 in Lapp et al. (Reference Lapp, Geraldo, Chien, Ay, Pakala, Batugedara, Humphrey, Ma, De, Le Roch, Galinski and Kissinger2017) and Giemsa-stained blood film showing mature schizont- and early trophozoite-infected erythrocytes.




