Introduction
To date, only 5 confirmed Plasmodium species are known to be transmitted from one human host to another by Anopheles mosquitoes, namely, Plasmodium vivax, Plasmodium falciparum, Plasmodium malariae, Plasmodium ovale wallikeri and Plasmodium ovale curtisi. Globally, malaria elimination efforts are centred on 2 prevalent parasites: P. falciparum is the most lethal, while P. vivax is the most widespread species (Battle et al., Reference Battle, Lucas, Nguyen, Howes, Nandi, Twohig, Pfeffer, Cameron, Rao, Casey, Gibson, Rozier, Dalrymple, Keddie, Collins, Harris, Guerra, Thorn, Bisanzio, Fullman, Huynh, Kulikoff, Kutz, Lopez, Mokdad, Naghavi, Nguyen, Shackelford, Vos, Wang, Lim, Murray, Price, Baird, Smith, Bhatt, Weiss, Hay and Gething2019; World Health Organization, 2022). Other human malaria Plasmodium species, like P. malariae infection, are prevalent throughout the tropics and subtropics. Plasmodium ovale curtisi and P. ovale wallikeri are sympatric sibling species commonly found in sub-Saharan Africa and Asia (Sutherland et al., Reference Sutherland, Tanomsing, Nolder, Oguike, Jennison, Pukrittayakamee, Dolecek, Hien, do Rosário, Arez, Pinto, Michon, Escalante, Nosten, Burke, Lee, Blaze, Otto, Barnwell, Pain, Williams, White, Day, Snounou, Lockhart, Chiodini, Imwong and Polley2010). The most common zoonotic malaria agent in Malaysia is Plasmodium knowlesi, the 6th species of human malarial parasite that was formerly believed to infect macaques but has since been found to infect humans (White, Reference White2008; Muhammad et al., Reference Muhammad, Azman, Eddie, Azmi, Yee and Idris2022; World Health Organization, 2022). Plasmodium knowlesi has also spread to some southeast Asian countries (White, Reference White2008; Muhammad et al., Reference Muhammad, Azman, Eddie, Azmi, Yee and Idris2022; World Health Organization, 2022). Although there have been no non-zoonotic malaria cases reported in Malaysia for the previous 4 years, there have been a total of 17 125 cases of P. knowlesi since 2017 (World Health Organization, 2022).
The evolution and origin of P. falciparum, the most virulent parasitic species in the Plasmodium genus, has been the subject of intense research and discussion for many decades. Recent reports suggest that both P. falciparum and P. vivax evolved from wild-living African apes, as demonstrated by whole genome sequencing (Loy et al., Reference Loy, Liu, Li, Learn, Plenderleith, Sundararaman, Sharp and Hahn2017). Plasmodium vivax originated from an ancestral stock of parasites that infected gorillas, chimpanzees and humans in Africa, while P. falciparum emerged from a cross-species parasite transmission from gorillas (Loy et al., Reference Loy, Liu, Li, Learn, Plenderleith, Sundararaman, Sharp and Hahn2017). A recent study illustrated that genome sequencing results of P. malariae that infects chimpanzees have similar selection characteristics to another Plasmodium lineage that can infect human and chimpanzee hosts (Fig. 1) (Rutledge et al., Reference Rutledge, Böhme, Sanders, Reid, Cotton, Maiga-Ascofare, Djimdé, Apinjoh, Amenga-Etego, Manske, Barnwell, Renaud, Ollomo, Prugnolle, Anstey, Auburn, Price, McCarthy, Kwiatkowski, Newbold, Berriman and Otto2017). A human P. ovale variant has also been discovered in African apes, demonstrating a natural cross-species exchange of P. ovale infections between chimpanzees and humans (Fig. 1) (Duval et al., Reference Duval, Nerrienet, Rousset, Sadeuh Mba, Houze, Fourment, Le Bras, Robert and Ariey2009; Rutledge et al., Reference Rutledge, Böhme, Sanders, Reid, Cotton, Maiga-Ascofare, Djimdé, Apinjoh, Amenga-Etego, Manske, Barnwell, Renaud, Ollomo, Prugnolle, Anstey, Auburn, Price, McCarthy, Kwiatkowski, Newbold, Berriman and Otto2017).

Figure 1. Non-human and human malaria primates considered in this study: erythrocytic cycle, their natural hosts, most common regions where the infections are reported from; similarities to other humans and their natural hosts. The details for natural hosts for Plasmodium species are adopted from Carlton (Reference Carlton2018) and Escalante and Pacheco (Reference Escalante and Pacheco2019).
Further, P. knowlesi infections are primarily zoonotic infections with wild macaques as their reservoir hosts, which could have adapted to switch their hosts to humans as the preferred hosts due to increasing human population and ecological changes (Fig. 1) (Lee et al., Reference Lee, Divis, Zakaria, Matusop, Julin, Conway, Cox-Singh and Singh2011). These studies suggest that malaria was a non-human primate (NHP) disease and that P. vivax and P. falciparum emerged as human infective agents in Africa with subsequent host-switching from gorillas (Loy et al., Reference Loy, Liu, Li, Learn, Plenderleith, Sundararaman, Sharp and Hahn2017). NHPs infecting Plasmodium species like Plasmodium cynomolgi, Plasmodium simium, Plasmodium inui, Plasmodium brasilianum, Plasmodium coatneyi and Plasmodium fieldi are among the zoonotic malaria species that have obtained the ability to infect humans via Anopheles (Sharp et al., Reference Sharp, Plenderleith and Hahn2020). In this study, we reveal trends in the emergence of NHP Plasmodium species and the global reports of their transmission to humans.
Materials and methods
Inclusion and exclusion criteria
We included only reports of NHP malaria infections in humans (either mono- or mixed-infections with any Plasmodium species) in humans with available full text. The exclusion criteria were: (1) studies including NHP Plasmodium species infection in their related natural hosts (Old and New World monkeys, chimpanzees and gorillas), (2) studies where sufficient reports could not be retrieved, (3) P. knowlesi infection reports in humans and (4) articles not available in English.
Information sources and search strategy
A systematic literature analysis was performed for all NHP malaria infections in humans. The data were collated and reviewed from the relevant literature that reports the cases of zoonotic Plasmodium infection transmission in humans from 2 search engines, PubMed and Medline. The terms used in the search were ‘rare Plasmodium species infecting humans’, ‘non-knowlesi zoonotic malaria’, ‘host switching’ and ‘macaque or non-human primate malaria’. Following this, we also used keywords for individual zoonotic ‘Plasmodium sp. (P. cynomolgi, P. brasilianum, P. inui, P. simium, P. coatneyi, and P. fieldi) human infections’, so no studies are missed for zoonotic human infections (Fig. 2). The time frame was not defined to check the initial reports of NHP infections in humans.

Figure 2. Flowchart depicting the study design for natural infections in humans by NHP Plasmodium species.
Study selection and data extraction
Two independent reviewers reviewed titles and abstracts to collect publications that matched the inclusion criteria. The entire text of the publications was retrieved and evaluated for eligibility if the title and abstract of the paper could not be rejected with certainty by both researchers. Tables 1 and 2 display the lists of NHP infection cases in humans.
Table 1. Details of Plasmodium cynomolgi infections in humans

a Accidental infection of B strain of P. cynomolgi in human.
b Experimental transmission of P. cynomolgi in humans.
Table 2. Details of Plasmodium inui, Plasmodium coatneyi, Plasmodium brasilianum, Plasmodium simium and Plasmodium fieldi infections in humans

Results
Study identification and selection
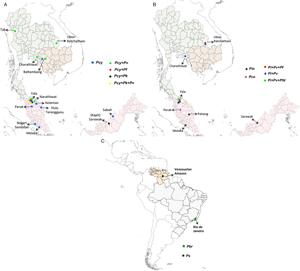
We considered only non-knowlesi Plasmodium species, as P. knowlesi infections are already known to infect humans and have emerged as the dominant species in Malaysia (Muhammad et al., Reference Muhammad, Azman, Eddie, Azmi, Yee and Idris2022). Nineteen studies from 377 search hits retrieved for NHP malaria infections in humans have been considered. Six NHP Plasmodium species with human transmission cases were individually included: P. brasilianum, P. coatneyi, P. cynomolgi, P. fieldi, P. inui and P. simium. Further, these 19 retrieved studies were also cross-checked by searching for individual studies for each Plasmodium zoonotic species, namely, P. brasilianum, P. coatneyi, P. cynomolgi, P. inui, P. fieldi and P. simium from 62, 62, 25, 233, 55 and 36 search hits, respectively. The spread of zoonotic species infections in humans is depicted in Fig. 3.

Figure 3. Locations of zoonotic Plasmodium species malaria infections in humans: (A) mono- and mixed-infections of Plasmodium cynomolgi, with Plasmodium falciparum, Plasmodium knowlesi and Plasmodium vivax in southeast Asian countries; (B) mono-infections of Plasmodium inui and Plasmodium coatneyi; mixed-infections of P. inui with P. falciparum, Plasmodium fieldi and P. vivax in southeast Asia and (C) mono-infections of Plasmodium brasilianum and Plasmodium simium in South America. The boundaries of countries are coloured as: Brazil (grey), Cambodia (light brown), Malaysia (pink), Thailand (light green) and Venezuela (nude). The shapefiles of the world map and the countries were downloaded from the University of Texas Libraries Geodata Portal (Hijmans, Reference Hijmans2019).
Geographical distribution of zoonotic species to humans
Plasmodium cynomolgi zoonosis
Plasmodium cynomolgi, the most recent NHP parasite infecting humans, was cultivated in vitro in the early 1980s (Nguyen-Dinh et al., Reference Nguyen-Dinh, Gardner, Campbell, Skinner and Collins1981). Plasmodium cynomolgi infections are predominantly found in macaque monkeys like Macaca fascicularis (long-tailed macaque) and Macaca nemestrina (pig-tailed macaque) (Fig. 1) (Chua et al., Reference Chua, Ong, Malleret, Suwanarusk, Kosaisavee, Zeeman, Cooper, Tan, Zhang, Tan, Abas, Yip, Elliot, Joyner, Cho, Breyer, Baran, Lange, Maher, Nosten, Bodenreider, Yeung, Mazier, Galinski, Dereuddre-Bosquet, Le Grand, Kocken, Rénia, Kyle, Diagana, Snounou, Russell and Bifani2019). However, P. cynomolgi infections are also reported in experimental and rare natural zoonotic infections in humans (Eyles et al., Reference Eyles, Coatney and Getz1960; Kuvin et al., Reference Kuvin, Beye, Stohlman, Contacos and Coatney1962; Bennett and Warren, Reference Bennett and Warren1965; Garnham, Reference Garnham1966). The first recorded P. cynomolgi infection was in M. fascicularis in Indonesia in 1907, which was later acquired naturally by humans throughout southeast Asia from various macaque monkeys (Coatney et al., Reference Coatney, Collins, Warren and Contacos1971; Kotepui et al., Reference Kotepui, Masangkay, Kotepui and Milanez2021). In terms of morphological and biological features, P. cynomolgi is nearly identical to its sister taxon P. vivax with its asexual replication, i.e. 48 h, the period between infection and the appearance of parasites in the blood (prepatent period), and the existence of a dormant stage (hypnozoites) (Fig. 1) (Cross et al., Reference Cross, Hsu-Kuo and Lien1973; Most, Reference Most1973; Druilhe et al., Reference Druilhe, Trape, Leroy, Godard and Gentilini1980). Both P. cynomolgi and P. vivax prefer to infect reticulocytes and have Schuffner's dot-modified infected erythrocyte membrane (Bykersma, Reference Bykersma2021). The experimental and accidental infection of humans with P. cynomolgi was shown half a century ago, with the suspicion that this simian parasite might infect humans and that an actual zoonotic outbreak would occur in the future (Cross et al., Reference Cross, Hsu-Kuo and Lien1973; Most, Reference Most1973; Druilhe et al., Reference Druilhe, Trape, Leroy, Godard and Gentilini1980).
The first accidental P. cynomolgi (P. cynomolgi bastianellii) infection in humans was reported in the Laboratory of Parasite Chemotherapy, National Institute of Allergy and Infectious Diseases, while studying the P. cynomolgi subspecies in rhesus monkeys (Macaca mulatta). Four accidental infections by this simian malaria occurred among laboratory workers, proving that one particular ‘B strain’ of P. cynomolgi could produce malaria in humans (Eyles et al., Reference Eyles, Coatney and Getz1960). The first naturally acquired P. cynomolgi human infection was reported in Hulu Terengganu, Peninsular Malaysia, in 2011, near a small forest crowded with macaques (Ta et al., Reference Ta, Hisam, Lanza, Jiram, Ismail and Rubio2014). The infection was initially diagnosed as P. malariae/P. knowlesi and later as P. vivax by microscopy and molecular methods. However, re-examination was performed via nested multiplex-polymerase chain reaction (PCR) followed by a parallel nested PCR for Plasmodium genus amplification, confirming the P. cynomolgi infection with asexual stages (Ta et al., Reference Ta, Hisam, Lanza, Jiram, Ismail and Rubio2014). In this region (Peninsular Malaysia), Anopheles cracens is the predominant mosquito species in Peninsular Malaysia and is hence suspected as a vector for the transmission of P. cynomolgi. A detailed survey conducted between 2011 and 2014 in 7 states of Malaysia identified 9 mono-infections of P. cynomolgi (8.8%) from multiple districts out of the 102 malaria-positive samples (Table 1) (Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). This study also reported the infections of other zoonotic Plasmodium species: P. coatneyi and P. inui, with ~3% prevalence of 102 malaria-positive individuals (Table 1) (Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). A malariometric study conducted in Cambodia between 2013 and 2016 reported that out of 1361 asymptomatic malaria-positive patients, 21 (~1.6%) were asymptomatic carriers of NHP malarial parasites (Imwong et al., Reference Imwong, Madmanee, Suwannasin, Kunasol, Peto, Tripura, von Seidlein, Nguon, Davoeung, Day, Dondorp and White2019). Of these 21 patients, 52.4% of asymptomatic patients had P. cynomolgi mono-infections, while ~9.6% carried mixed-infections of P. cynomolgi and P. vivax (Imwong et al., Reference Imwong, Madmanee, Suwannasin, Kunasol, Peto, Tripura, von Seidlein, Nguon, Davoeung, Day, Dondorp and White2019). From 2013 to 2017, a study of 1047 malaria-positive patients in Kapit, Malaysia, reported mixed-infections of P. cynomolgi and P. knowlesi in 6 clinical cases (~0.6%) in Sarawak, Malaysian Borneo (Raja et al., Reference Raja, Hu, Kadir, Mohamad, Rosli, Wong, Hii, Simon Divis and Singh2020). Another study conducted in 2015 in Sabah, Malaysia, identified 2 asymptomatic P. cynomolgi mono-infections (3.07%) out of 54 malaria-positive cases (Grignard et al., Reference Grignard, Shah, Chua, William, Drakeley and Fornace2019). In Thailand during 2007–2018, out of 1180 symptomatic malaria patients reported via species-specific nested PCR, 9 were P. cynomolgi (0.76% prevalence) co-infections with P. vivax (0.59%), P. falciparum (0.09%) and P. vivax + P. knowlesi (0.09%) (Table 1) (Putaporntip et al., Reference Putaporntip, Kuamsab, Pattanawong, Yanmanee, Seethamchai and Jongwutiwes2021). Most P. cynomolgi cases were reported in areas where macaques were in close proximity to humans (wild or domesticated). The study could not determine if P. cynomolgi caused symptomatic infections or coexisted asymptomatically with other human malarial parasites (Putaporntip et al., Reference Putaporntip, Kuamsab, Pattanawong, Yanmanee, Seethamchai and Jongwutiwes2021).
Owing to the high prevalence of nocturnal mosquitoes and macaques, Thailand and similar regions may be a potential infection source for P. cynomolgi transmission to humans. In 2018, a traveller with a P. cynomolgi symptomatic infection was reported to have visited Thailand and Peninsular Malaysia (Hartmeyer et al., Reference Hartmeyer, Stensvold, Fabricius, Marmolin, Hoegh, Nielsen, Kemp and Vestergaard2019). Another study conducted in Thailand in 2021 to probe simian Plasmodium species in blood samples of malaria patients reported 21 mono-infections with P. cynomolgi (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). These 2 reports from Thailand highlight the occurrence of P. cynomolgi in diverse malaria-endemic areas of Thailand (Hartmeyer et al., Reference Hartmeyer, Stensvold, Fabricius, Marmolin, Hoegh, Nielsen, Kemp and Vestergaard2019; Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). Further, Yala province in Thailand reported the highest number of mixed-infections (n = 17) of P. cynomolgi with other Plasmodium species (Fig. 3A) (Putaporntip et al., Reference Putaporntip, Kuamsab, Pattanawong, Yanmanee, Seethamchai and Jongwutiwes2021, Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). A recent systematic survey between 1946 and 2020 in southeast Asian countries compared the prevalence of P. cynomolgi infections in humans, mosquitoes and macaques in natural settings (Kotepui et al., Reference Kotepui, Masangkay, Kotepui and Milanez2021). The study demonstrated that the pooled proportion of naturally acquired P. cynomolgi was highest in macaques (47%), followed by mosquitoes (18%) and humans (1%) (Kotepui et al., Reference Kotepui, Masangkay, Kotepui and Milanez2021). Given this, P. cynomolgi transmission from mosquitoes to humans is likely constrained by the presence of macaque and Anopheles bite rates with their susceptibility (Kotepui et al., Reference Kotepui, Masangkay, Kotepui and Milanez2021).
Plasmodium inui zoonosis
Plasmodium inui, a quartan-type parasite, infects Old World monkeys with a typical 72 h quartan periodicity. Plasmodium inui natural hosts are macaques (Macaca nigra, Macaca cyclopis, M. fascicularis, M. mulatta, M. nemestrina, and Macaca radiate), and the primate infections are reported throughout Asia, including southern India, southeast Asia and Taiwan (Fig. 1; Table 2) (Eyles, Reference Eyles1963; Coatney et al., Reference Coatney, Collins, Warren and Contacos1971). One vector responsible for P. inui transmission – Anopheles leucosphyrus (a vector of human malaria in Sarawak, Borneo) was identified in 1962, which was caught while biting a man, showing the possibility of transmission of a monkey infection to humans in nature (Coatney et al., Reference Coatney, Collins, Warren and Contacos1971). Following this, 2 volunteers established the experimental natural transmission of P. inui to man via bites of infected mosquitoes (Anopheles stephensi/Anopheles maculatus) (Coatney et al., Reference Coatney, Chin, Contacos and King1966). In 2020, an epidemiologic and entomological study from Pahang, Malaysia, revealed 2 natural, asymptomatic mono-infections of P. inui by nested PCR (Liew et al., Reference Liew, Bukhari, Jeyaprakasam, Phang, Vythilingam and Lau2021). All the tested individuals were participants who underwent forest training in 2020. The primers aimed at asexual and sexual 18S rRNA genes confirmed infection with P. inui (Liew et al., Reference Liew, Bukhari, Jeyaprakasam, Phang, Vythilingam and Lau2021). Anopheles cracens and An. leucosphyrus were shown to be the possible vector of monkey infection and transmission to humans in the wild. Both cases experienced minimal symptoms, and the parasitaemia was undetectable for short periods. Hence, the quartan P. inui parasite could be self-limiting in humans since it was not detected approximately 8 months after a patient's exposure to an infectious mosquito bite (Liew et al., Reference Liew, Bukhari, Jeyaprakasam, Phang, Vythilingam and Lau2021). According to a surveillance study, there was a 66.7% predominance of P. inui in all macaques tested in Pahang (26/39 macaques sampled), suggesting that humans may get infected with P. inui via vector-borne transmission from infected macaques to humans (Amir et al., Reference Amir, Shahari, Liew, de Silva, Khan, Lai, Snounou, Abdullah, Gani, Rovie-Ryan and Lau2020; Liew et al., Reference Liew, Bukhari, Jeyaprakasam, Phang, Vythilingam and Lau2021). In 2021, blood samples from malaria patients in 5 malaria-endemic regions of Thailand confirmed the natural transmission of P. inui in 19 patients out of all reported human and other infections (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). Most patients infected with P. inui had concurrent infections with other Plasmodium species (Fig. 3B) (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). Further, in 2011 and 2014, the blood samples from 14 villages in 7 states in Malaysia of indigenous populations of various sub-tribes recorded 3 P. inui infections while searching for P. cynomolgi cases (Table 2) (Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). However, another study conducted from 2014 to 2015 from communities in Sarawak, Malaysian Borneo regions did not detect any P. inui and P. cynomolgi infections in humans (Siner et al., Reference Siner, Liew, Kadir, Mohamad, Thomas, Zulkarnaen and Singh2017). The locations of reported mono- and mixed-infections of P. inui in humans are shown in Fig. 3B.
Plasmodium simium zoonosis
Plasmodium simium was first identified in 1951 in the blood smear of Alouatta clamitans (brown howler monkeys) collected in the Atlantic Forest in Brazil (Fonseca, Reference Fonseca1951; Deane et al., Reference Deane, Deane and Ferreira Neto1966). Plasmodium simium is found in south Brazil, where it is found among woolly spider monkeys (Brachyteles arachnoids), capuchin monkeys (Cebus and Sapajus sp.), arboreal howler monkeys (Alouatta sp.) and black-fronted titi monkeys (Callicebus nigrifrons) (Fig. 1) (Coatney et al., Reference Coatney, Collins, Warren and Contacos1971). The first suspected natural infection of a human by P. simium was from Brazil (Deane et al., Reference Deane, Deane and Ferreira Neto1966). The infection was suspected on the basis that it had occurred in a forest reserve outside São Paulo where P. simium was known to be transmitted along with the morphological characteristics of the parasite (Deane et al., Reference Deane, Deane and Ferreira Neto1966). After half a decade, in 2015–2016, an epidemiological investigation of malaria patients in Rio de Janeiro, Brazil, reported a total of 28 mono-infections of P. simium out of 49 autochthonous malaria cases indicative of P. simium zoonosis in Brazil, although initially misdiagnosed as P. vivax (Fig. 3C; Table 2) (Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017). The mitochondrial genome of these cases indicated that P. simium was most closely related to the South American P. vivax parasite (Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017). The P. simium sequence revealed that it is similar to P. vivax, corroborating earlier claims that it originated from a host switch from humans to monkeys. Further, a study identified 7 distinct haplotypes from 22 human blood isolates (infected with P. vivax identified by microscopy) from Atlantic Forest inhabitants in Espírito Santo, Brazil (Fig. 3C). Of these 7 isolates, 2 isolates when shared with samples obtained from simians had an identical sequence to P. simium (Table 2) (Buery et al., Reference Buery, Rodrigues, Natal, Salla, Loss, Vicente, Rezende, Duarte, Fux, Malafronte, Falqueto and Cerutti2017). This suggests that P. simium has been endemic in the Atlantic region but may have been incorrectly diagnosed as P. vivax due to the lack of any reliable diagnostic molecular techniques (Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017; Buery et al., Reference Buery, Rodrigues, Natal, Salla, Loss, Vicente, Rezende, Duarte, Fux, Malafronte, Falqueto and Cerutti2017). Plasmodium simium then adapted to the monkeys, and now occasionally infects humans in the region due to opportunistic infections (de Oliveira et al., Reference de Oliveira, Rodrigues, Early, Duarte, Buery, Bueno, Catão-Dias, Cerutti, Rona, Neafsey and Ferreira2021; Mourier et al., Reference Mourier, de Alvarenga, Kaushik, de Pina-Costa, Douvropoulou, Guan, Guzmán-Vega, Forrester, de Abreu, Júnior, de Souza Junior, Moreira, Hirano, Pissinatti, Ferreira-da-Cruz, de Oliveira, Arold, Jeffares, Brasil, de Brito, Culleton, Daniel-Ribeiro and Pain2021). The primary vector in this area is suspected to be the Anopheles (kerteszia) cruzi which is found almost exclusively in the Atlantic region and can feed on both monkeys in the canopy and humans at the ground level (Deane et al., Reference Deane, Deane and Ferreira Neto1966, 196; Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017). The isolates of P. simium from the New World monkeys and humans have a close genome-wide association with P. vivax from the New World. The genome-wide divergence between P. simium and New World P. vivax is negligible compared to intraspecific polymorphism in P. vivax populations of South America (de Oliveira et al., Reference de Oliveira, Rodrigues, Early, Duarte, Buery, Bueno, Catão-Dias, Cerutti, Rona, Neafsey and Ferreira2021). The differences between P. vivax and P. simium are focused on large deletions in the P. simium Duffy-binding protein 1 and reticulocyte-binding protein 2a genes which are usually present in all human-derived isolates (Mourier et al., Reference Mourier, de Alvarenga, Kaushik, de Pina-Costa, Douvropoulou, Guan, Guzmán-Vega, Forrester, de Abreu, Júnior, de Souza Junior, Moreira, Hirano, Pissinatti, Ferreira-da-Cruz, de Oliveira, Arold, Jeffares, Brasil, de Brito, Culleton, Daniel-Ribeiro and Pain2021). There are only 2 unique single-nucleotide polymorphisms (SNPs) in the P. simium mitochondrial genome, differentiating it from P. vivax (Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017; Rodrigues et al., Reference Rodrigues, Valdivia, De Oliveira, Alves, Duarte, Cerutti-Junior, Buery, Brito, De Souza, Hirano, Malafronte, Ladeia-Andrade, Mita, Santamaria, Calzada, Kawamoto, Raijmakers, Mueller, Pacheco, Escalante, Felger and Ferreira2018). Further, a recent study also identified 8 P. simium-specific SNPs out of 9 infected humans using an inexpensive tool specific to diagnose P. simium infections (De Alvarenga et al., Reference De Alvarenga, Culleton, De Pina-Costa, Rodrigues, Bianco, Silva, Nunes, De Souza, Hirano, Moreira, Pissinatti, De Abreu, Lisboa Areas, Lourenço-de-Oliveira, Zalis, Ferreira-da-Cruz, Brasil, Daniel-Ribeiro and De Brito2018). Hence, it can be speculated that gene deletions in human-derived isolates and other genetic changes in the P. simium genome may have helped to invade human red blood cells (RBCs), thus explaining the basis of recent zoonotic infections (de Oliveira et al., Reference de Oliveira, Rodrigues, Early, Duarte, Buery, Bueno, Catão-Dias, Cerutti, Rona, Neafsey and Ferreira2021; Mourier et al., Reference Mourier, de Alvarenga, Kaushik, de Pina-Costa, Douvropoulou, Guan, Guzmán-Vega, Forrester, de Abreu, Júnior, de Souza Junior, Moreira, Hirano, Pissinatti, Ferreira-da-Cruz, de Oliveira, Arold, Jeffares, Brasil, de Brito, Culleton, Daniel-Ribeiro and Pain2021).
Plasmodium brasilianum zoonosis
In New World monkeys, P. brasilianum is a simian parasite that causes quartan fever (Fig. 1) (Contacos et al., Reference Contacos, Lunn, Coatney, Kilpatrick and Jones1963). Plasmodium brasilianum was identified in at least 35 species of New World primates in Central and South America (Chaves et al., Reference Chaves, Dolz, Ibarra-Cerdeña, Núñez, Ortiz-Malavasi, Bernal-Valle and Gutiérrez-Espeleta2022). The genetic and morphological characteristics of P. brasilianum are indistinguishable from those of P. malariae (Contacos et al., Reference Contacos, Lunn, Coatney, Kilpatrick and Jones1963; Fandeur et al., Reference Fandeur, Volney, Peneau and De Thoisy2000). Thus, P. brasilianum and P. malariae may specialize in different hosts but remain members of the same quartan malarial species. This anthropozoonotic parasite can easily circulate between humans or NHPs (Contacos et al., Reference Contacos, Lunn, Coatney, Kilpatrick and Jones1963). Investigations in the 1960s supported the theory that P. brasilianum could experimentally infect humans from monkeys and vice versa (Coatney et al., Reference Coatney, Collins, Warren and Contacos1971). A comparison of the circumsporozoite protein and ribosomal small subunit (18S) in parasites of 75 P. malariae-positive patients revealed 16% with parasites that were nearly identical to the strain of P. brasilianum of infected monkeys from French Guiana (Fandeur et al., Reference Fandeur, Volney, Peneau and De Thoisy2000; Lalremruata et al., Reference Lalremruata, Magris, Vivas-Martínez, Koehler, Esen, Kempaiah, Jeyaraj, Perkins, Mordmüller and Metzger2015). These studies indicate that P. brasilianum is endemic to Latin America and that P. brasilianum and P. malariae parasites can spread quickly between humans and monkeys, serving as a natural reservoir for malaria (Lalremruata et al., Reference Lalremruata, Magris, Vivas-Martínez, Koehler, Esen, Kempaiah, Jeyaraj, Perkins, Mordmüller and Metzger2015). The presence of P. brasilianum has already been established in howler monkeys in Central America (Costa Rica) (Chinchilla et al., Reference Chinchilla, Guerrero, Gutiérrez and Sánchez2006). Interestingly, a group identified 3 samples with 99% identity with P. malariae/P. brasilianum from human clinical samples in Costa Rica (Fig. 3C; Table 2) (Calvo et al., Reference Calvo, Morera, Dolz, Solorzano-Morales and Herrero2015). The analysis revealed a 99% identity with P. malariae isolated from atypical human cases in Asia, and a 99% identity with a sequence of P. brasilianum isolated from a non-human monkey of Guiana (Calvo et al., Reference Calvo, Morera, Dolz, Solorzano-Morales and Herrero2015). Similarly, another study demonstrated the genomic sequence identity of 99.70% in mitochondrial and apicoplast genomes of P. brasilianum with P. malariae (Talundzic et al., Reference Talundzic, Ravishankar, Nayak, Patel, Olsen, Sheth, Batra, Loparev, Vannberg, Udhayakumar and Barnwell2017). Additionally, it is established that while belonging to the radiation of human P. malariae strains, P. brasilianum does not represent a separate lineage and that P. brasilianum likely emerged after the human infection was transmitted to New World monkeys (Plenderleith et al., Reference Plenderleith, Liu, Li, Loy, Mollison, Connell, Ayouba, Esteban, Peeters, Sanz, Morgan, Wolfe, Ulrich, Sachse, Calvignac-Spencer, Leendertz, Shaw, Hahn and Sharp2022).
Plasmodium fieldi zoonosis
Plasmodium fieldi asexual cycle is 48 h (Fig. 1). Bonnet macaques (Macaca radiata), long-tailed macaques (M. fascicularis), baboons (Papio doguera), rhesus macaques (M. mulatta) and pig-tailed macaques (M. nemestrina) are noted as natural hosts and reservoirs of P. fieldi (Eyles, Reference Eyles1963). In 2021, P. fieldi was reported to be capable of cross-transmission between macaques and humans under natural conditions (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). While examining the symptomatic malaria patients in Thailand, a P. fieldi infection was diagnosed in 3 out of 5271 tested patients (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). All P. fieldi-infected patients had concurrent infections with other Plasmodium species and responded well to chloroquine or artemisinin–mefloquine combination therapy (Table 2) (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). A study conducted in Thailand determining the prevalence of different Plasmodium species in NHPs reported that of 93 macaque blood samples examined, P. inui (35%) and P. fieldi (30%) were the most prevalent species in malaria-positive macaques, presenting them as the natural reservoir and a potential public health concern to the local population (Fungfuang et al., Reference Fungfuang, Udom, Tongthainan, Kadir and Singh2020). The geographical distribution of reported P. fieldi infections in humans is depicted in Fig. 3B.
Plasmodium coatneyi zoonosis
Plasmodium coatneyi is commonly found in long-tailed macaques (M. fascicularis), and unlike other simian parasites, P. coatneyi shares morphological features to P. falciparum (Fig. 1) (Eyles, Reference Eyles1963; Fungfuang et al., Reference Fungfuang, Udom, Tongthainan, Kadir and Singh2020). A detailed study conducted between 2011 and 2014 in 7 states of Malaysia showed 3 mono-infections (2.17%) of P. coatneyi among 645 samples that tested positive for malaria (Table 2) (Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). This study highlighted the existence of naturally acquired human infection with P. coatneyi, a species earlier believed to be incapable of infecting humans through infected monkey blood or mosquito bites (Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). The geographical spread of P. coatneyi in humans is shown in Fig. 3B.
Potential basis for transmission and their control
NHPs in their ‘forest habitat’, the Leucosphyrus-group Anopheles vectors, and human proximity to NHP habitats are linked to zoonotic malaria transmission. The ecology of mosquitoes' reservoir hosts and vectors is an important factor influencing the spread of zoonotic malaria to humans (van de Straat et al., Reference van de Straat, Sebayang, Grigg, Staunton, Garjito, Vythilingam, Russell and Burkot2022). In southeast Asia, the main species transmitting NHP malarial parasites and human-only malaria species in a few regions belong predominantly to the An. leucosphyrus (Moyes et al., Reference Moyes, Henry, Golding, Huang, Singh, Baird, Newton, Huffman, Duda, Drakeley, Elyazar, Anstey, Chen, Zommers, Bhatt, Gething and Hay2014). From the perspective of public health, the Leucosphyrus subgroup is reported to be an effective vector, as numerous species have been implicated as zoonotic malaria vectors, including those for P. knowlesi (Collins et al., Reference Collins, Contacos and Guinn1967, Reference Collins, Contacos, Skinner and Guinn1971). Traditional classifications of the Anopheles dirus and An. leucosphyrus complex species are considered forest and forest fringe inhabitants (Faust and Dobson, Reference Faust and Dobson2015). The interactions between humans, mosquitoes and monkeys, all of which are heterogeneous in space and time, are influenced by each group's behaviour. Infected mosquitoes must bite humans for zoonotic Plasmodium species to infect them. This necessitates being close to an infectious vector, which is often linked to shifts in land use, occupation and housing design (Ramasamy, Reference Ramasamy2014; Johnson et al., Reference Johnson, Kumar Sharma, Cuenca, Byrne, Salgado-Lynn, Shahar, Lin, Zulkifli, Mohd Saidi, Drakeley, Matthiopoulos, Nelli and Fornace2022). Additionally, humans and vectors that readily prey on humans and reservoir animals must be in proximity to the reservoir hosts or wildlife that harbours parasites. In their natural hosts, macaques, P. knowlesi and P. cynomolgi typically cause benign, long-lasting infections (Anderios et al., Reference Anderios, NoorRain and Vythilingam2010). As a result, since the illness does not affect the monkeys' natural behaviours, infected monkeys make the best reservoirs for transmitting parasites to humans (Antinori et al., Reference Antinori, Galimberti, Milazzo and Corbellino2013).
The increasing reports of zoonotic/NHP Plasmodium species infecting humans within a decade are alarming. There might be multiple reasons for malaria transmission from primates to human by Anopheles. These might be (1) a growing need for more land for humans with the rapid increase in the human population in some regions that overlap with zoonotic infections; (2) substantial deforestation in tropical malaria-endemic countries (Kar et al., Reference Kar, Kumar, Singh, Carlton and Nanda2014); (3) increased contact between humans and mosquitoes that feed on NHPs along with increased interactions between humans and macaques due to urbanization and encroachment of NHP habitats (Kar et al., Reference Kar, Kumar, Singh, Carlton and Nanda2014); (4) the sequence analysis of NHP Plasmodium species underpins the genetic adaptations in the simian parasite that allow invasion of human RBCs and may explain the basis of recent zoonotic to human infections (Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017) and (5) entomological factors such as vector distribution and potential, parasite prevalence and environmental patterns may also contribute to human transmission (Mills et al., Reference Mills, Gage and Khan2010; Kar et al., Reference Kar, Kumar, Singh, Carlton and Nanda2014).
The use of indoor residual spraying and long-lasting insecticide-impregnated bed nets are 2 current mitigation strategies for reducing zoonotic malaria (World Health Organization, 2022). Nevertheless, these strategies might prove insufficient for other zoonotic control, as seen in P. knowlesi infections (Scott, Reference Scott2020), since these strategies neglect the parasite's ongoing transmission between the host-animal reservoir populations.
There is no strong evidence for the efficacy of current antimalarial drugs against NHP parasite infections, but some studies reported the resolution of clinical symptoms with antimalarial drugs. Plasmodium cynomolgi infection was treated with atovaquone + proguanil followed by primaquine in the European traveller (Hartmeyer et al., Reference Hartmeyer, Stensvold, Fabricius, Marmolin, Hoegh, Nielsen, Kemp and Vestergaard2019), artemether + lumefantrine followed by primaquine in Malaysian patients, and chloroquine + primaquine or artesunate + mefloquine in Thailand patients. Primaquine is prescribed as the primary antimalarial drug treatment because of P. cynomolgi parasite relapse; this must be made aware to the clinicians. Hence, the treatment of P. cynomolgi malaria shows a significant knowledge gap. A study in Thailand showed that P. inui and P. fieldi were responsive to chloroquine or artemisinin–mefloquine treatment (Putaporntip et al., Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022). In Brazil, P. simium-infected patients responded successfully to chloroquine and primaquine without hospital admission or relapses (Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017).
Discussion
Several studies conducted over half a century ago suggested that simian malaria could be transmitted to humans in experimental and accidental settings (Coatney et al., Reference Coatney, Chin, Contacos and King1966; Deane et al., Reference Deane, Deane and Ferreira Neto1966; Cross et al., Reference Cross, Hsu-Kuo and Lien1973; Most, Reference Most1973; Druilhe et al., Reference Druilhe, Trape, Leroy, Godard and Gentilini1980; Lalremruata et al., Reference Lalremruata, Magris, Vivas-Martínez, Koehler, Esen, Kempaiah, Jeyaraj, Perkins, Mordmüller and Metzger2015). However, natural transmission of these (P. cynomolgi, P. brasilianum, P. inui, P. simium, P. coatneyi and P. fieldi) primate-related parasites to humans has been reported from 2010 onwards (Deane et al., Reference Deane, Deane and Ferreira Neto1966; Ta et al., Reference Ta, Hisam, Lanza, Jiram, Ismail and Rubio2014; Lalremruata et al., Reference Lalremruata, Magris, Vivas-Martínez, Koehler, Esen, Kempaiah, Jeyaraj, Perkins, Mordmüller and Metzger2015; Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017; Grignard et al., Reference Grignard, Shah, Chua, William, Drakeley and Fornace2019; Hartmeyer et al., Reference Hartmeyer, Stensvold, Fabricius, Marmolin, Hoegh, Nielsen, Kemp and Vestergaard2019; Imwong et al., Reference Imwong, Madmanee, Suwannasin, Kunasol, Peto, Tripura, von Seidlein, Nguon, Davoeung, Day, Dondorp and White2019, 201; Raja et al., Reference Raja, Hu, Kadir, Mohamad, Rosli, Wong, Hii, Simon Divis and Singh2020; Liew et al., Reference Liew, Bukhari, Jeyaprakasam, Phang, Vythilingam and Lau2021; Putaporntip et al., Reference Putaporntip, Kuamsab, Pattanawong, Yanmanee, Seethamchai and Jongwutiwes2021, Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022; Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). Zoonotic malaria has been reported in humans in many parts of southeast Asia under natural conditions through the bite of infected Anopheles mosquitoes (Deane et al., Reference Deane, Deane and Ferreira Neto1966; Ta et al., Reference Ta, Hisam, Lanza, Jiram, Ismail and Rubio2014; Lalremruata et al., Reference Lalremruata, Magris, Vivas-Martínez, Koehler, Esen, Kempaiah, Jeyaraj, Perkins, Mordmüller and Metzger2015; Brasil et al., Reference Brasil, Zalis, De Pina-Costa, Siqueira, Júnior, Silva, Areas, Pelajo-Machado, De Alvarenga, Da Silva Santelli, Albuquerque, Cravo, Santos De Abreu, Peterka, Zanini, Suárez Mutis, Pissinatti, Lourenço-de-Oliveira, De Brito, De Fátima Ferreira-da-Cruz, Culleton and Daniel-Ribeiro2017; Grignard et al., Reference Grignard, Shah, Chua, William, Drakeley and Fornace2019; Hartmeyer et al., Reference Hartmeyer, Stensvold, Fabricius, Marmolin, Hoegh, Nielsen, Kemp and Vestergaard2019; Imwong et al., Reference Imwong, Madmanee, Suwannasin, Kunasol, Peto, Tripura, von Seidlein, Nguon, Davoeung, Day, Dondorp and White2019, 201; Raja et al., Reference Raja, Hu, Kadir, Mohamad, Rosli, Wong, Hii, Simon Divis and Singh2020; Liew et al., Reference Liew, Bukhari, Jeyaprakasam, Phang, Vythilingam and Lau2021; Putaporntip et al., Reference Putaporntip, Kuamsab, Pattanawong, Yanmanee, Seethamchai and Jongwutiwes2021, Reference Putaporntip, Kuamsab, Seethamchai, Pattanawong, Rojrung, Yanmanee, Weng Cheng and Jongwutiwes2022; Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). Because of their proximity to monkey reservoir hosts and mosquito vectors, people who live on the forest periphery, work in or travel in forested areas are most likely to get zoonotic malaria (Kar et al., Reference Kar, Kumar, Singh, Carlton and Nanda2014; Lalremruata et al., Reference Lalremruata, Magris, Vivas-Martínez, Koehler, Esen, Kempaiah, Jeyaraj, Perkins, Mordmüller and Metzger2015; Kotepui et al., Reference Kotepui, Masangkay, Kotepui and Milanez2021). The absence of highly sensitive techniques for detecting the parasites to differentiate between morphological and genetic similarities was a significant hurdle in correctly identifying Plasmodium species. To identify and confirm Plasmodium species, all reports included in the current study used either highly sensitive nested PCR or semi-nested for small subunit rRNA gene or cytochrome C oxidase gene amplification and sequencing. It is inferred that P. cynomolgi, P. fieldi, P. simium and P. inui infections are zoonotic transmissions that possibly originated and diverged from P. vivax while infections of P. brasilianum are from P. malariae (Sharp et al., Reference Sharp, Plenderleith and Hahn2020). Based on the cumulative studies, zoonotic malaria transmission has only occurred in South America and southeast Asian countries until now (Fig. 3). A close examination of epidemiology and parasite transmission also revealed that the distribution of zoonotic malaria cases depends entirely on Plasmodium species and on the demographics of the human host populace in different geographical locations (van de Straat et al., Reference van de Straat, Sebayang, Grigg, Staunton, Garjito, Vythilingam, Russell and Burkot2022). For example, most P. cynomolgi, P. inui, P. fieldi and P. coatneyi cases are reported in Asian countries, while P. simium and P. brasilianum cases are from South America (Fig. 3). Compared to other zoonotic cases, the proportion of mono-/mixed-infections caused by P. cynomolgi was more prevalent than other reported rare infections. The P. cynomolgi infections were reported from southeast Asian countries, majorly from Thailand, and it is alarming that the number of reported P. cynomolgi cases has increased in the past 10 years (2011–2020). In contrast, P. simium and P. brasilianum are endemic in South America (Brazil and Venezuela) (Fig. 3). Unlike other simian parasites, P. fieldi cases have been reported recently in 2021 from Thailand, mostly, and all P. fieldi co-exist with other malarial parasite species only (Imwong et al., Reference Imwong, Madmanee, Suwannasin, Kunasol, Peto, Tripura, von Seidlein, Nguon, Davoeung, Day, Dondorp and White2019).
In contrast, P. inui infections in humans are widespread throughout southern Asia (Imwong et al., Reference Imwong, Madmanee, Suwannasin, Kunasol, Peto, Tripura, von Seidlein, Nguon, Davoeung, Day, Dondorp and White2019; Yap et al., Reference Yap, Hossain, Nada-Raja, Ngui, Muslim, Hoh, Khaw, Kadir, Simon Divis, Vythilingam, Singh and Lim2021). No mixed-infections have been reported with P. simium, P. brasilianum and P. coatneyi until now. So, unlike P. cynomolgi and P. simium, the rarely reported cases of P. fieldi, P. inui and P. coatneyi require additional future research efforts. The establishment and spread of these zoonotic species as human-infecting Plasmodium species highlight the importance of understanding how parasites' transmission capabilities adapt to new hosts, and to predict future zoonotic malaria outbreaks. It is clear that NHP malarial parasites are a potential reservoir of infectious human parasites. A molecular assessment of these infections is provided by malariometric studies of asymptomatic human infections with NHP parasites. These surveys typically use microscopy, rapid diagnostic tests or non-species-specific PCR, making it difficult to identify malarial parasites that infect NHPs. In this situation, whole-genome amplification and species-specific PCR will be beneficial. More surveillance studies and possible control measures should be considered to curtail the transmission of these parasites to achieve malaria elimination worldwide. Further, the increased encroachment of reservoir hosts by humans and the outdoor blood-biting habits of the vectors also pose challenges to mitigation efforts due to zoonotic malaria (Scott, Reference Scott2020). Hence, a trans-disciplinary approach targeting vectors' contact with humans must be considered rather than the conventional malaria control methods. This can be accentuated by monitoring wild macaques and their carriers. It would be advisable to understand better the epidemiology of the Plasmodium parasites they harbour and develop effective strategies for minimizing the potential threat of zoonotic malaria infections.
Data availability statement
All data are included in the paper.
Author's contribution
R. C.: data acquisition, analysis and interpretation, drafting the manuscript and editing the manuscript critically. S. B.: data collection and analysis and drafting the manuscript, K. B.: data acquisition and analysis. A. S.: Original idea, conceptually designing the work, data interpretation, drafting the manuscript and revising it critically.
Financial support
This study is not supported by any funding source.
Competing interests
None.