Introduction
Tomato leaf curl New Delhi virus (ToLCNDV) has become a significant constraint in melon-growing regions worldwide, causing severe crop damage. It is a bipartite begomovirus with DNA A and DNA B and is spread in a persistent manner by the whitefly (Bemisia tabaci). ToLCNDV was first reported on tomato (Solanum lycopersicum L.) in India (Papidam et al., Reference Papidam, Beachy and Fouquet1995; Srivastava et al., Reference Srivastava, Hallan, Raizada, Chandra, Singh and Sane1995; Mandal, Reference Mandal, Sharma, Gaur and Ikegami2010; Varma et al., Reference Varma, Mandal, Singh and Thompson2011). Later, it was found in other South and Southeast Asian nations. The host range was expanded to include other plant species, mainly crops from the Cucurbitaceae and Solanaceae families (Chang et al., Reference Chang, Ku, Tsai, Chien and Jan2010; Pratap et al., Reference Pratap, Kashikar and Mukherjee2011; Jyothsna et al., Reference Jyothsna, Haq, Singh, Sumiya, Praveen, Rawat, Briddon and Malathi2013). Prior to 2012, ToLCNDV was exclusively reported in Asian nations. Subsequently, it spread westward, with documented cases in the Middle East, Spain, Portugal, Italy, Greece, Morocco, Tunisia, and Algeria within the Mediterranean Basin (Juarez et al., Reference Juarez, Tovar, Fiallo-Olive, Aranda, Gonsalves, Castillo, Moriones and Navas-Castillo2014; Mnari-Hattab et al., Reference Mnari-Hattab, Zammouri, Belkadhi, Bellon Doña, ben Nahia and Hajlaoui2015; Ruiz et al., Reference Ruiz, Simón, Velasco, García and Janssen2015; Panno et al., Reference Panno, Lacono, Davino, Marchione, Zappardo, Bella, Tomassoli, Accotto and Davino2016; Sifres et al., Reference Sifres, Sáez, Ferriol, Selmani, Riado, Pico and López2018; Kheireddine et al., Reference Kheireddine, Sifres, Sáez, Pico and López2019; Orfanidou et al., Reference Orfanidou, Malandraki, Beris, Kektsidou, Vassilakos, Varveri, Katis and Maliogka2019; EPPO, 2020). Since the initial report, ToLCNDV outbreaks have significantly impacted cucurbit production in the Andalusia region of southern and southeastern Spain, particularly in Almeria, Murcia and Malaga (Juárez et al., Reference Juarez, Tovar, Fiallo-Olive, Aranda, Gonsalves, Castillo, Moriones and Navas-Castillo2014, Reference Juárez, Rabadán, Martínez, Tayahi, Grande-Pérez and Gómez2019). Almeria, hosting the world's largest concentration of greenhouses, faces potential economic losses in the production of zucchini (6358 ha), watermelon (5665 ha), cucumber (4920 ha) and melon (3740 ha) (Valera et al., Reference Valera, Belmonte, Molina-Aiz and López2016) due to ToLCNDV infection. The European and Mediterranean Plant Protection Organization (EPPO) added this virus to the EPPO Alert List due to its transmission potential (EPPO, 2020). ToLCNDV poses a significant challenge for both open-field and greenhouse-grown cucurbits. Sequence analysis of the strain spreading in the Mediterranean territory revealed a new isolate named ToLCNDV-ES, originating from the recombination of ToLCNDV strains. ToLCNDV-ES, better adapted to cucurbits than its Indian relative, poses a threat to European cucurbit production (Fortes et al., Reference Fortes, Sánchez-Campos, Fiallo-Olivé, Díaz-Pendón, Navas-Castillo and Moriones2016).
The most common ToLCNDV symptoms on melon plants include yellow spots, yellow mosaic, smaller leaves, short internodes, leaf curling, thicker leaf margins, darkening, puckering and severe stunting of the entire plant (Padmanabha et al., Reference Padmanabha, Choudhary, Mishra, Mandal, Solanke, Mishra and Yadav2023). Early stage infections cause severe curling and puckering in young leaves, significantly reducing growth and total abortion of fruits. This virus affects plants at all stages of development and usually leads to significant crop loss (Wilisiani et al., Reference Wilisiani, Neriya, Tagami, Kaneko, Hartono, Nishigawa and Natsuaki2019).
The muskmelon (Cucumis melo L.; 2n = 2x = 24) is an old world warm-season cucurbit species, a member of the Cucurbitaceae family. Muskmelons are cherished for their appealing fruit, which has a distinct aromatic musky flavour and a sweet taste. They are considered a healthy food (Munshi and Choudhary, Reference Munshi, Choudhary, Peter and Hazra2014) as they contain abundant ascorbic acid, carotene, folic acid, potassium and other bioactive components beneficial to human health. Due to its higher genetic diversity, C. melo is recognized as one of the most varied and polymorphic species in the Cucurbitaceae family. This species comprises six horticultural groupings based on the type and uses of the fruit: cantalupensis (cantaloupe and muskmelon), inodorus (winter melon), flexuosus (long melon), conomon (pickling melon), dudaim (pomegranate melon) and momordica (snap melon). In addition to being enjoyed as a delicious fruit, the leaves and seeds of muskmelons are also used to treat hematomas. This further highlights the numerous benefits and applications of this versatile and beneficial fruit.
The transient nature of whitefly outbreaks and the insect vectors resistance to different pesticides pose fresh challenges to the control of viral diseases (Caballero et al., Reference Caballero, Cyman and Schuster2013). Viral recombination and mutation can lead to the creation of new virus strains and species, enabling them to adapt to changing environmental factors and weakening the resistance mechanisms (Mansoor et al., Reference Mansoor, Zafar and Briddon2006).
Islam et al. (Reference Islam, Munshi, Mandal, Kumar and Behera2010) found that the genotypes DSG-6 and DSG-7 of sponge gourd were resistant to ToLCNDV infection in both natural epiphytotic and challenge inoculation screening methods. Lopez et al. (Reference Lopez, Ferriol and Pico2015) identified three C. melo subsp. agrestis var. momordica (Kharbuja, PI 124112, PI 414723) and two wild agrestis (WM-7 and WM-9) accessions of melon with ToLCNDV tolerance. These accessions showed either no symptoms or moderate symptoms, tested negative for PCR and had low virus load. PI 604506 and PI 381814, two ToLCNDV-resistant Cucurbita moschata lines were discovered by Saez et al. (Reference Saez, Martinez, Ferriol, Manzano, Velasco, Jamilena, Lopez and Pico2016), which exhibited very low virus titres. Masegosa et al. (Reference Masegosa, Martinez, Aguado, Garcia, Cebrian, Iglesias-Moya, Paris and Jamilena2020) found that the Japanese-origin C. moschata accession, BSUAL-252, was highly resistant to ToLCNDV. This accession showed no symptoms after whitefly-mediated and mechanical inoculation methods. Saez et al. (Reference Saez, Ambrosio, Miguel, Valcarcel, Diez, Pio and Lopez2021) identified three Indian cucumber accessions (CGN23089, CGN23423 and CGN23633) that were extremely resistant to ToLCNDV.
Given the challenges of managing viral diseases, resistance breeding is the only economically and ecologically viable way to reduce disease incidence. Williams (Reference Williams1980) proposed genetic resistance as a successful strategy to combat these diseases, as it defines the best breeding techniques for disease resistance programmes. Romay et al. (Reference Romay, Pitrat, Lecoq, Wipf-Scheibel, Millot, Girardot and Desbiez2019) reported that resistance to ToLCNDV in the melon accession ‘IC-274014’ is controlled by a single recessive gene. In contrast, Saez et al. (Reference Saez, Esteras, Martínez, Ferriol, Dhillon, Lopez and Pico2017) reported single dominance of inheritance for ToLCNDV in the melon accession WM-7 (C. melo. subsp. agrestis var. agrestis) and epistatic interaction was also reported in another source of resistance DSM 132 (Padmanabha et al., Reference Padmanabha, Choudhary, Mishra, Mandal, Solanke, Mishra and Yadav2023) in a different cross by our group. These contrasting reports on the inheritance of ToLCNDV in melons suggested that genetics of ToLCNDV are determined by the source of resistance used and different sources with varied genetic mechanism will always be useful in the crop improvement programme.
Saez et al. (Reference Saez, Esteras, Martínez, Ferriol, Dhillon, Lopez and Pico2017) identified the SNP marker D16, which is closely linked to the ToLCNDV resistance gene on chromosome 11 in melon. In cucumber, the closest marker SNPCS2_3 is linked to the ToLCNDV resistance quantitative trait locus (QTL) located on chromosome 02 (Saez et al., Reference Saez, Ambrosio, Miguel, Valcarcel, Diez, Pio and Lopez2021). A major QTL controlling resistance to ToLCNDV was identified in pumpkin on chromosome 8. The interval of the four QTLs was flanked by the DPM34 and D133 markers (Saez et al., Reference Saez, Martínez, Montero-Pau, Esteras, Sifres, Blanca, Ferriol, Lopez and Pico2020).
The development of molecular markers linked to disease resistance genes has made considerable progress in resistance breeding. These markers can identify novel resistant sources without the need for pathogen inoculation, thereby reducing the duration of breeding programmes. In gene pyramiding programmes, molecular markers have been proved as an effective tool. It facilitates the introduction of different resistance genes into a single cultivated line, enhancing disease resistance effectiveness and durability, consequently reducing the cost of breeding resistant plants (Slater et al., Reference Slater, Cogan and Forster2013). Cleaved amplified polymorphic sequence (CAPS), a PCR-based marker system, is capable of distinguishing between resistant and susceptible genotypes, and its findings complement the phenotypic data (Panthee and Chen, Reference Panthee and Chen2009).
The development of the first powdery mildew-resistant melon cultivar ‘PMR 45’, released in California, utilized Snapmelon (C. melo var. momordica) germplasm. Line PI 79376 was used in the development of ‘PMR 5’ in response to the emergence of Podosphaera xanthii race 2 (Whitaker and Davis, Reference Whitaker and Davis1962). The contemporary varieties of muskmelon resistant to race 2 of P. xanthii and to Golovinomyces cichoracearum trace their origins back to this genetic stock. Following this, additional Indian snapmelon accessions, namely PI 124111, PI 124112, PI 134192 and PI 414723, demonstrated resistance to various diseases and pests, including downy mildew, Fusarium wilt, zucchini yellow mosaic virus, papaya ringspot virus, cucurbit aphid-borne yellow virus and Aphis gossypii (Pitrat et al., Reference Pitrat, Hanelt and Hammer2000). Indian muskmelon cultivars have incorporated resistance to cucumber green mottle mosaic virus from Indian snapmelon (Pan and More, Reference Pan and More1996). Furthermore, sources of resistance from snapmelon germplasm (DSM-11) have been identified at IARI, New Delhi, which could be utilized for the development of a resistant genotype of muskmelon against Fusarium wilt disease (Choudhary et al., Reference Choudhary, Dhar and Kalia2013).
The primary centre of diversity for the cultivated melon is considered to be India, according to recent taxonomic and molecular research findings. Indian germplasm is genetically distinct from African germplasm, which makes independent domestication events possible (Sebastian et al., Reference Sebastian, Schaefer, Telford and Renner2010). Higher levels of resistance to various plant diseases are found in wild relatives or landraces of other cultivated species. In this context, we have identified line DSM-19 (C. melo var. momordica) as a novel source of resistance to ToLCNDV form Indian melon germplasm at the Indian Agricultural Research Institute (IARI), New Delhi after 2 years of natural epiphytotic screening in 2019 and 2020, and the resistance in this line has also been validated through challenge inoculation by using viruliferous whiteflies in 2020 (Padmanabha et al., Reference Padmanabha, Choudhary, Mishra, Mandal, Solanke, Mishra and Yadav2023). To our knowledge, information on the genetics of ToLCNDV resistance in melon is very scanty in India. Taking all of these factors into account, we attempted to report on the inheritance pattern of ToLCNDV disease associated with melon using the wild germplasm line DSM-19 as a novel source of resistance and also identified a CAPS marker linked with ToLCNDV resistance.
Materials and methods
Experimental site
The experiment was conducted at the Division of Vegetable Science, IARI in New Delhi, India. The farm is situated at an elevation of approximately 228 m above mean sea level, at 20° 40′ North latitude and 77°13′ East longitude. The climate in this region is subtropical, with an average temperature ranging from 25 to 32°C. The soil at the farm is loamy sand, black and rich in organic matter.
Development of populations for inheritance study
During July–October 2019 and 2020, the resistant source DSM-19 (C. melo var. momordica) underwent evaluation against ToLCNDV disease in natural epiphytotic conditions for 2 years. In 2020, the same line was revalidated through challenge inoculation, demonstrating consistent resistance in both screening methods (Padmanabha et al., Reference Padmanabha, Choudhary, Mishra, Mandal, Solanke, Mishra and Yadav2023). To study the inheritance of ToLCNDV, a susceptible genotype called Pusa Sarda (C. melo var. inodorus) was crossed with DSM-19 to generate F1s. The F1 seeds were used to grow plants, which were then selfed by wrapping the flowers in cotton wads. Mature seeds from self-fruits were collected and sowed to produce the F2 generation. Additionally, BC1P1 and BC1P2 populations were created by backcrossing F1 plants with Pusa Sarda and DSM-19, respectively.
Screening under natural epiphytotic conditions
ICAR-IARI, New Delhi, which comes under Trans-Gangetic ecological region of northern India, is a hotspot for ToLCNDV disease in cucurbits due to its unique location and climatic conditions during the Kharif season (July–November). During both years of natural epiphytotic screening at the experimental farms of the Division of Vegetable Science, IARI, New Delhi, the whitefly population started to increase from July to August during Kharif season and reached its highest peak during October–November. These trends are mainly due to climatic factors, where the combination of high temperature and humidity results in a sharp increase in the whitefly population due to its rapid multiplication. Additionally, the large population of whiteflies, attracted by the cultivation of a broad range of vegetables such as tomato, brinjal, chillies and various cucurbits, contributes to the prevalence of the major pathogen ToLCNDV, which is primarily transmitted by these insect vectors. Under irrigated field conditions, screening for ToLCNDV resistance was conducted using a standardized six-point interaction system phenotype scale (Sohrab, Reference Sohrab2005). Thirty plants each of parent 1, parent 2 and F1 (Pusa Sarda × DSM-19) were assessed, along with 152 plants of the F2 population and 60 plants each of BC1P1 and BC1P2. The scale used for scoring was as follows: 0 = no symptoms (immune, I); 1 = mild mosaic pattern in young leaves covering <10% area (resistant, R); 2 = mosaic pattern in young leaves covering <25% area (moderately resistant, MR); 3 = mosaic pattern in young leaves covering <50% area, blistering and puckering of leaves (moderately susceptible, MS); 4 = widespread mosaic pattern in young leaves covering <75% area, distortion of leaves (susceptible, S); 5 = widespread mosaic pattern in young leaves covering >75% area, distortion of leaves and stunting of the plants. Eight scorings were taken at weekly intervals after the appearance of the first disease symptoms on the susceptible line (20 days after sowing). To ensure successful cultivation of the crop, all plants were sown at a spacing of 120 cm × 45 cm, and recommended cultural practices were followed, with the exception of insecticide application.
Screening through challenge inoculation
Seeds of parental lines, together with F1 and F2 generations (generated by self-pollination of the F1), and backcross generations consisting of 30 plants of each parent, 30 plants of F1, 140 plants of F2 and 40 plants of each BC1P1 and BC1P2 of the melon cross Pusa Sarda × DSM-19 were sown in plastic pots of 7.50 × 6.0 cm size. Whitefly colonies were reared and maintained on healthy tobacco plants under controlled conditions in insect-rearing cages. An ideal whitefly population was maintained by providing a temperature of 28–35°C, a relative humidity of 30–50%, and a 14 h photoperiod. To make the whiteflies viruliferous for screening, they were allowed to feed on ToLCNDV-infected muskmelon plants. After 24 h of feeding (acquisition access period) on the infected muskmelon plants, the whiteflies were considered virulent and used for challenge inoculation of 15-day-old healthy seedlings (at the 2–4 true leaf stages) of all six generations for 24 h. For screening, each plant was individually inoculated, and 10 viruliferous whiteflies were released per plant. Plant evaluation was performed using a standardized six-point interaction system phenotype scale (Sohrab, Reference Sohrab2005), as explained in the section above (screening under natural epiphytotic conditions). Disease scoring on each plant was conducted from 7 days post-inoculation (dpi) and continued at weekly intervals for up to 2 months post-inoculation.
Design and development of CAPS marker for ToLCNDV resistance
The CAPS 16 (2) marker was developed based on the SNP marker D16, located on chromosome 11 of melon. This SNP marker was reported to be associated with resistance to ToLCNDV, representing a major QTL (Saez et al., Reference Saez, Esteras, Martínez, Ferriol, Dhillon, Lopez and Pico2017). To design the CAPS 16 (2) marker, the nucleotide sequence of SNP D16 was blasted in NCBI, revealing a 100% similarity match with the C. melo genomic chromosome, chr_11 (sequence ID: LN713265.1; length: 31442130), within the range of 29690253–29690799. The restriction enzyme site GGCC for the enzyme Hae III was not found at the earlier reported D16 SNP location [T/A] but we could identify the restriction site GGCC at 220 bp away from D16 SNP [T/A] on the same gene. Hence, the primers were designed to capture the GGCC restriction site which is cleaved by the Hae III enzyme which leads to polymorphism. Using this sequence, a 546 bp PCR product (including a BsuRI [Hae III] enzyme cleavage site ‘GGCC’) was amplified with the forward primer CAPSF: 5′AACATCTGTCAGTTGGTGAAA3′ and the reverse primer CAPSR: 5′CCCACTGAAACGTCAACCC3′.
Genome pool construction and DNA extraction
Based on the results of challenge inoculation of the Pusa Sarda × DSM-19 F2 population, two genomic DNA pools were constructed: R-bulk (resistant to ToLCNDV) and S-bulk (susceptible to ToLCNDV). These pools were created by mixing an equal concentration of DNA from 10 ToLCNDV-resistant (score-1) and 10 ToLCNDV-susceptible (score-5) F2 individuals. CAPS marker identification was performed using these two bulks and their parents. Apical leaves were collected from inoculated plants 60 dpi. Total genomic DNA was isolated using the cetyl trimethyl ammonium bromide method (Murray and Thompson, Reference Murray and Thompson1980; Doyle and Doyle, Reference Doyle and Doyle1990). The DNA was then quantified using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and diluted with sterile deionized water to achieve a final concentration of 100 ng/l.
PCR amplification and restriction digestion
To ensure parental polymorphism, PCR amplification of the resistant parent DSM-19 and susceptible parent Pusa Sarda was carried out using the CAPS 16 (2) primer followed by restriction digestion. PCR amplification of the two parent DNA was carried out in a total volume of 20 μl. Each reaction mixture contained 100 ng of template DNA (1 μl), 10 μM of each specific forward and reverse primers (1 μl each), 10 μl of 2× the master mix (Gene DireX, Inc., Taiwan) and 7 μl of sterile deionized water. The PCR was performed using the Eppendorf Master Thermo Cycler with the following cycling conditions: an initial denaturation at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 45 s and extension at 72°C for 1.0 min. A final extension step was carried out at 72°C for 10 min. Subsequently, restriction digestion of 10 μl of the amplified products was performed overnight at 37°C in a total volume of 20 μl. The digestion mixture included 7 μl of distilled water, 2 μl of the restriction enzyme 10× buffer and 1 μl of BsuRI (Hae III) restriction enzyme (10 U/μl) from Thermo Fisher Scientific. The digested products were analysed by agarose gel electrophoresis using a 2% (w/v) gel and visualized using ethidium bromide staining. Once the parental polymorphism was confirmed, PCR amplification and restriction digestion of the two parent DNA samples and two bulk samples (resistant bulk and susceptible bulk) were carried out as per the above-stated procedure.
Sequencing of polymorphic PCR products
The PCR amplification products obtained from Pusa Sarda and DSM-19 were subjected to sequencing in order to confirm PCR product digestion with a specific restriction enzyme. The forward and reverse sequences were then aligned using the CAP 3 DNA sequence assembly programme to generate contigs. Subsequently, the contigs from both resistant and susceptible parents, along with the reference genome, were aligned using the Clustal Omega software (http://www.ebi.ac.uk/Tools/msa/clustalo). The obtained results were analysed for further insights.
Statistical analysis
Investigation of the expression of ToLCNDV disease in all six generations resulting from the cross between Pusa Sarda × DSM-19 was conducted under both natural epiphytotic screening and challenge inoculation. The hypothesis of monogenic control of ToLCNDV resistance was tested using a χ2 test for goodness of fit (Panse and Sukhatme, Reference Panse and Sukhatme1985). A probability (P) value greater than 0.05 was considered appropriate for the genetic model. Analysis was performed by using SAS 9.3 software package available at Indian Agricultural Statistics Research Institute, New Delhi, India.
Results
Natural epiphytotic and challenge inoculation screening
To understand the Mendelian inheritance of ToLCNDV resistance under natural epiphytotic screening and challenge inoculation conditions, we classified individual plants based on their disease symptoms. Plants scoring ‘0’, ‘1’ and ‘2’ (i.e. those with <25% symptom coverage on their leaves) were considered resistant, while those scoring ‘3’, ‘4’ and ‘5’ (i.e. >25% symptom coverage on leaves) were classified as susceptible. During the natural epiphytotic screening, all 30 DSM-19 plants exhibited resistance, while all 30 Pusa Sarda plants were found to be susceptible. In the F1 hybrid generation, all 30 plants were susceptible, indicating that susceptibility is dominant. In the F2 generation, out of 152 plants, 32 were resistant and 120 were susceptible, following a classic 1:3 Mendelian segregation pattern. For the BC1P1 generation, all 60 plants fell into the susceptible category. In contrast, for the BC1P2 generation, out of 60 plants, 35 were resistant and 25 were susceptible (Table 1).
Table 1. Estimates of χ2 values and their probability for classical Mendelian ratio for ToLCNDV resistance in the cross of Pusa Sarda × DSM-19 under natural epiphytotic conditions

χ2 at 0.05 with 1 df = 3.84.
Under challenge inoculation, all the DSM-19 showed resistance response, and all the Pusa Sarda plants showed a susceptible response. The population of 140 F2 plants showing various degree of symptoms was segregated into 29 resistant and 111 susceptible plants (Fig. 1), with the F1 plants being susceptible. BC1P1 population of 40 plants was all susceptible, whereas the 40 plants in the BC1P2 population were segregated into 16 resistant and 24 susceptible plants (Table 2).

Figure 1. ToLCNDV symptoms observed on F2 plants derived from Pusa Sarda × DSM-19 under challenge inoculation: (a) resistant F2 plants, (b) susceptible F2 plants.
Table 2. Estimates of χ2 values and their probability for classical Mendelian ratio for ToLCNDV resistance in the cross of Pusa Sarda × DSM-19 under challenge inoculation
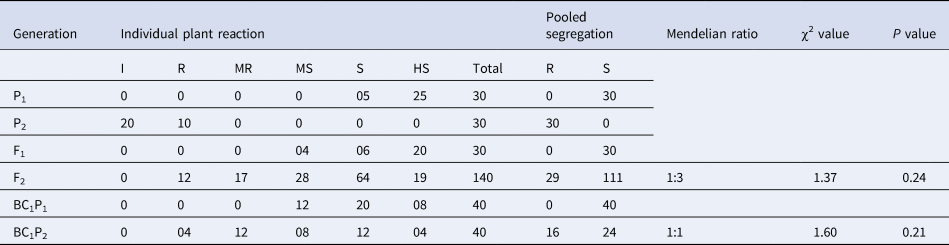
χ2 at 0.05 with 1 df = 3.84.
Genetics of resistance
Upon natural screening and challenge inoculation, it was discovered that F1 individuals were susceptible, indicating that the resistant gene exhibits a recessive trait. Under natural epiphytotic conditions, the computed χ2 value was 1.26, whereas under challenge inoculation, it was 1.37 in F2 populations. In both screening procedures, the observed χ2 value for BC1P1 was 0, while for BC1P2, the χ2 values under natural epiphytotic and challenge inoculation were 1.67 and 1.60, respectively. These values were lower than table χ2 value of 3.84. The observed and expected frequencies are in close agreement, as calculated χ2 values are non-significant (Tables 1 and 2).
CAPS marker linked to ToLCNDV resistance
The PCR amplification of DNA was performed using Primer CAPS 16 (2) with susceptible parent Pusa Sarda and resistant parent DSM-19, resulting in a 546 bp amplicon size band. The advantage of using this primer lies in the potential to subject the amplicons to restriction enzyme digestion, which allows for the observation of polymorphism that characterizes the susceptible and resistant accessions. Upon digestion with restriction enzyme BsuRI (Fig. 2), the PCR product of the susceptible parent Pusa Sarda displayed two distinct bands with molecular sizes of 456 and 90 bp, indicating the presence of the BsuRI restriction site on this genotype. In contrast, the resistant accession DSM-19 showed an undigested fragment of 546 bp, indicating the absence of the BsuRI site. Primer CAPS 16 (2) effectively differentiated between the susceptible parent Pusa Sarda and the resistant parent DSM-19 (Fig. 2), making it a valuable tool for distinguishing between these two genotypes.

Figure 2. Amplicons generated from DNA isolated from two parents using the CAPS 16 (2) marker and were cleaved using BsuRI. M, 50 bp DNA ladder; SP, susceptible parent Pusa Sarda; RP, resistant parent DSM-19.1.
CAPS 16 (2) primer amplified 543 bp sequenced reads of susceptible parent Pusa Sarda and 530 bp of resistant parent DSM-19 aligned to a NCBI reference sequence: LN 713265.1 and length 31442130 (range: 29690253–29690799) on C. melo genomic Chr-11 containing particular SNP. Nucleotide base variation due to transition of A–G was observed in susceptible genotype Pusa Sarda; it is a site for the restriction enzyme, which distinguishes the resistant and susceptible genotype (Fig. 3).

Figure 3. Multiple sequence alignments of amplified region of CAPS16 (2) primer between susceptible genotypes Pusa Sarda and resistant genotype DSM-19 using the clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo). SNPs are highlighted with colour backgrounds.
After amplification with CAPS 16 (2) primer, PCR products of two parents and two bulks were subjected to digestion with the restriction enzyme BsuRI. This enzyme cleaves the DNA from the susceptible parent and susceptible bulk into two fragments, measuring 456 and 90 bp, respectively. In contrast, the DNA from the resistant parent and resistant bulk remains uncleaved (Fig. 4).

Figure 4. Amplicons were generated from DNA isolated from plants in two parents and two bulks using the CAPS 16 (2) marker and were cleaved using BsuRI. M, 50 bp DNA ladder; SP, susceptible parent Pusa Sarda; RP, resistant parent DSM-19; SB, susceptible bulk generated from extreme susceptible plants of F2 population of Pusa Sarda × DSM-19; RB, resistant bulk generated from extreme resistant plants.
Discussion
In breeding programmes aimed at transferring genes from one species to another, genetic background plays a crucial role in determining plant virus resistance. DSM-19 has been proven to be resistant under both natural epiphytotic screening and challenge inoculation. The F2 population confirmed the recessive nature of resistance as resistant and susceptible plants segregated, with the susceptible plants out numbering the resistant ones. The observed segregation pattern in the F2 population was consistent with the 1:3 ratio expected for monogenic recessive resistance. To further investigate the resistance locus linked to ToLCNDV resistance in muskmelon, a backcross population (BC1P1) was produced by crossing with susceptible parents. Surprisingly, every plant in the BC1P1 population turned out to be completely susceptible. However, in the subsequent backcross population (BC1P2), the plants segregated into susceptible and resistant individuals at a 1:1 ratio, confirming the monogenic recessive nature of resistance in DSM-19. These findings align with previous research on the genetic regulation of ToLCNDV resistance in cucurbits. For instance, Kaur et al. (Reference Kaur, Varalakshmi, Kumar, Rao, Pitchaimuthu, Mahesha, Venugopalan and Reddy2021) demonstrated the recessive nature of resistance genes to ToLCNDV in sponge gourd through two crosses of Arka Prasan × IIHR-Sel-1 and Arka Prasan × IIHR 137. In Cucurbita moschata and Cucumis sativus, a major recessive gene was found controlling resistance to ToLCNDV in an Indian accession (Saez et al., Reference Saez, Martínez, Montero-Pau, Esteras, Sifres, Blanca, Ferriol, Lopez and Pico2020, Reference Saez, Ambrosio, Miguel, Valcarcel, Diez, Pio and Lopez2021). Conversely, resistance to ToLCNDV in melon was reported to be controlled by a major QTL (Saez et al., Reference Saez, Esteras, Martínez, Ferriol, Dhillon, Lopez and Pico2017) and involving at least two major complementary dominant genes (Padmanabha et al., Reference Padmanabha, Choudhary, Mishra, Mandal, Solanke, Mishra and Yadav2022) in different source of resistance DSM-132. Additionally, in the case of sponge gourd (L. cylindrica), resistance to ToLCNDV is controlled by a single dominant gene (Islam et al., Reference Islam, Munshi, Mandal, Kumar and Behera2010). It is evident that the diverse nature of resistance genetics can be attributed to the different resistant sources being evaluated and the specific virus against which resistance is being tested.
In our inheritance study for ToLCNDV resistance, we used the resistant parent DSM-19 and observed that both the resistant bulk and the resistant parent had an uncleaved amplicon size of 546 bp after restriction with the enzyme. This suggests that the ToLCNDV resistance gene originates from DSM-19 and exhibits a nearly complete recessive pattern. The susceptible parent Pusa Sarda and susceptible bulk showed cleavage of the amplicon product obtained through CAPS marker 16 (2) into two fragments. This susceptibility might be attributed to a transition of SNP nucleotide from A to G. CAPS marker 16 (2) was designed near to the D16 SNP marker linked to the ToLCNDV resistance QTL located on chromosome 11 of melon (Saez et al., Reference Saez, Esteras, Martínez, Ferriol, Dhillon, Lopez and Pico2017). This resistant QTL region was derived from an Indian wild melon accession belonging to the agrestis group, similar to the DSM-19 used in our study, which belongs to subsp. agrestis var. momordica group. Furthermore, the BLAST analysis of SNP D16 sequence in NCBI predicted a gene called histidine kinase 3 with 85% similarity. According to literature citations, histidine kinase provides resistance against bacterial and fungal infections (Nongpiur et al., Reference Nongpiur, Soni, Karan, Singla-Pareek and Pareek2012).
In addition, a protein similar to the serine/threonine-protein kinase PBS1 of A. thaliana (MELO3C022340, located at chromosome 11 position 29,959,231–29,964,198) is annotated in this region. PBS1 was found to be highly upregulated in tomato cultivars that were resistant to ToLCNDV and is required for the plant defence mechanism mediated by R proteins (Sahu et al., Reference Sahu, Rai, Chakraborty, Singh, Chandrappa, Ramesh, Chattopadhyay and Prasad2010; Saez et al., Reference Saez, Esteras, Martínez, Ferriol, Dhillon, Lopez and Pico2017). This finding proves that the CAPS 16 (2) marker, designed from the major QTL located on chromosome 11 of melon, is linked to ToLCNDV resistance. There might also be sequences associated with eukaryotic translation initiation factors (Yeam, Reference Yeam2016) or messenger RNA surveillance factors, such as Pelota (found in locus ty-5), which confers resistance to geminiviruses in tomato. These findings provide evidence of the identified CAPS 16 (2) marker being linked to ToLCNDV resistance gene on chromosome 11 of melon. Similar results were observed in watermelon by Ling et al. (Reference Ling, Harris, Meyer, Levi, Guner, Wexner and Havey2008), where they identified a CAPS marker in an elF4E gene linked to zucchini yellow mosaic virus resistance, and its inheritance pattern was recessive. The identified CAPS 16 (2) marker linked to the ToLCNDV resistance gene in this study will be validated for transferability across various populations in future studies before using it in the screening of ToLCNDV-resistant accessions.
Conclusion
The information on ToLCNDV genetics in muskmelon germplasm in India and even globally is very scanty. This is the first report of monogenic recessive ToLCNDV resistance in a resistant source found through challenged inoculation, to the best of our knowledge. DSM-19 and cultivated muskmelon are cross-compatible, which means that the ToLCNDV resistance trait can be easily transferred to cultivated species via simple backcross breeding. This provides an excellent opportunity to develop high-yielding ToLCNDV-resistant muskmelon varieties. Furthermore, the CAPS 16 (2) marker, linked to the ToLCNDV-resistant gene, can be utilized to screen C. melo genotypes for resistance to ToLCNDV, once it is validated in a larger population.
Acknowledgments
The authors acknowledged the ICAR-Indian Agricultural Research Institute (IARI) for providing financial support to this study.
Author contributions
K. Padmanabha: methodology, management of crop, data recording and compilation, photography, writing and original draft. H. Choudhary: conceptualization, resources, supervision, review and editing. Gyan Mishra: supervision, review and editing. Bikash Mandal: conceptualization, resources, review and editing. Amolkumar Solanke: resources, review and editing. Dwij C. Mishra: data analysis and editing. Ramesh Kumar Yadav: supervision, review and editing.
Competing interests
None.








