Introduction
Cryoconite holes are variously shaped, water filled depressions that are distributed over the glaciers of polar, alpine and other mountainous areas of the world. These holes are the biologically active niches within glacial ecosystems (Säwström and others Reference Säwström, Mumford and Marshall2002). These contain soft, dark coloured granular material, mostly consisting of both organic and inorganic matter. The organic matter mainly includes algae, bacteria, (Takeuchi and others Reference Takeuchi, Kohshima and Seko2001; Säwström and others Reference Säwström, Mumford and Marshall2002; Anesio and others Reference Anesio, Hodson and Fritz2009; Kastovska and others Reference Kastovska, Elster, Stibal and Santruckova2005; Hodson and others Reference Hodson, Anesio and Tranter2008; Edwards and others Reference Edwards, Anesio and Rassner2011; Singh and others Reference Singh, Singh and Dhakephalkar2013a) and rotifers, while the inorganic matter is a mixture of minerals and trace elements (Singh and others Reference Singh, Sharma and Gawas2012). Recently, Singh and Singh (Reference Singh and Singh2012) characterised yeast and filamentous fungi from cryoconite holes of Midre Lovénbreen glacier, Svalbard. Further, and subsequently, a novel species Rhodotorula svalbardensis was reported from cryconites (Singh and others Reference Singh, Singh and Tsuji2014). Edwards and others (Reference Edwards, Douglas and Anesio2013) analysed fungi from cryoconites of Austre Brøggerbreen (AB), Midre Lovénbreen (ML) and Vestre Brøggerbreen (VB) glaciers with terminal-restriction fragment length polymorphism (T-RFLP) profiles, and recorded the presence of Articulospora and Varicosporium through a cultured approach. However, studies on fungal community inhabiting cryoconite holes on glaciers in Svalbard are still scanty.
Cryoconite holes work as mico-ecosystems and have ecological and biotechnological importance. To date there has been no study on the enzyme producing ability of cryophilic fungi from cryoconites of Brøggerbreen glaciers of Svalbard, Arctic. This study aims to address this knowledge gap by characterising the fungi inhabiting these glaciers cryoconites.
Materials and methods
Study area and sample collection
Samples were collected from Brøggerbreen glaciers (Austre and Vestre) (Fig 1a). Cryoconite samples were collected from different locations of glaciers in ablation zone. Austre Brøggerbreen (11.7 km2) and Vestre Brøggerbreen (5.3 km2) glaciers are situated on the western part of Spitsbergen, Svalbard. These two glaciers are the main sources of water to the Bayelva river (also known as Red river) finally merging into Kongsfjorden. Cryoconites were collected into sampling bags following strict contamination-free procedures (using a sterile gloves, syringe and sterile HiMedia sample collector), transported to laboratory with dry ice and stored at -20°C until processed.

Fig. 1. a) Map showing the sampling areas, b) Landscape of Vestre Brøggerbreen glacier in Svalbard Arctic, c) Cryoconite holes d) protease activity e) lipase activity f) cellulase activity g) amylase activity h) urease activity.
Isolation of yeast and filamentous fungi
One gram of cryoconite sediment was processed following the serial dilution method (Waksman Reference Waksman1916) and plated on mycological media MEA (malt extract agar, pH 5.5), PDA (potato dextrose agar, pH 5.6 ± 0.2), SDA (Sabouraud dextrose agar, pH 5.6) and PCA (potato carrot agar, pH 6.8 ± 0.2) (HiMedia India), by pour plate as well as spread plate techniques. Plates were incubated in triplicates at 4, 15, 22 and 25ºC for 2–4 weeks. Culture plates were monitored regularly and on the basis of shape, colour, and different morphological features (hyphae, conidiophore, and conidial structure). The distinct colony was picked up, sub cultured, and observed for purity of cultures under a microscope. The purified fungal colonies were transferred onto PDA slants (in a test tube at about a 35º slant to provide more surface area for fungal growth), stored at 4ºC for detailed study.
For morpho-taxonomical studies, the fungal mounts were prepared on slides using lactophenol-cotton blue as a mounting medium, and observed under Olympus BX-51 and IX-71 model microscopes. Fungal cultures were initially identified on the basis of morphotaxonomy with the help of standard literatures (Ellis Reference Ellis1971, Reference Ellis1976; Barron Reference Barron1977; Carmichael and others Reference Carmichael, BryceKendrick, Connersand and Sigler1980; Kirk and others Reference Kirk, Cannon and Minter2008; De Hoog and others Reference De Hoog, Gottlich and Platas2005; Kurtzman and others Reference Kurtzman, Fell and Boekhout2011). The isolates with similar morphological characteristics were grouped together, and the representative isolates were subjected to DNA sequence analysis. All identified pure cultures were maintained on PDA slants and deposited at the National Fungal Culture Collection of India (NFCCI-WDCM 932) in Pune, India.
Molecular characterisation (polymerase chain reaction (pcr), sequencing and phylogenetic analysis)
Total DNA was extracted from cultures (grown on PDA for 3 weeks at 4ºC) using the ISOPLANT II kit (Wako pure chemical industries Ltd.,Japan). Extracted DNA was amplified by PCR method using KOD-plus DNA polymerase (Toyobo Co. Ltd. Japan). PCR was performed under following conditions: Initial denaturation at 95°C for 2 min followed by 35 cycles of each denaturation at 95°C for 1 min, annealing at 52°C for 30 sec, elongation at 72°C for 1 min and final elongation was carried out at 72°C for 7 min.
The ITS region was amplified using following primers: ITS1F (5’-GTA ACA AGG TTT CCG T) and ITS4 (5´-TCC TCC GCT TAT TGA TAT GC). The amplified DNA was purified using a Wizard® SV Gel and PCR Clean-Up System (Promega KK, Tokyo, Japan). The purified DNA was sequenced on ABI DNA Sequencer following standard protocol.
The sequences of isolates were analysed using the NCBI database and BLAST. Sequence alignment of ITS region isolates, together with the homologous sequences (retrieved from Genbank) of closely related species, was performed using Clustal W option of MEGA software version 6.0 (Tamura and others Reference Tamura, Stecher and Peterson2013). Subsequently the DNA sequences of the four isolates (AB703291, AB703292, KT223585 and KT223586) were deposited in the data bank.
To calculate the sequence divergence, the matrix was analysed using the neighbour joining method (Saitou and Nei Reference Saitou and Nei1987), the Tamura-Nei model (Tamura and Nei Reference Tamura and Nei1993) and the Maximum Parsimony method (Tamura and others Reference Tamura, Peterson and Peterson2011). To represent the evolutionary history of the taxa, the bootstrap consensus tree was inferred from 1000 replicates (Felsenstein Reference Felsenstein1985). The pairwise alignment was performed using EMBOSS Matcher - Pairwise Sequence Alignment tool (www.ebi.ac.uk/Tools/psa/emboss_matcher/nucleotide.html).
Screening for enzymatic activity
The enzyme activity (amylase, cellulase lipase and protease) was determined at 1, 4, 10, and 20˚C according to established procedures (Hankin and Anagnostakis Reference Hankin and Anagnostakis1975; Buzzini and Martini Reference Buzzini and Martini2002). Urease activity was also tested on YNBG (6.7 g/l Yeast Nitrogen Base, 20g/l glucose) containing 1 g/l urea solution (pH 5.5). The diameter of the clear zone was measured to apparently quantify the enzyme activity (Figs 1d-g). Change in color from orange to pink was considered positive (Kurtzman and others Reference Kurtzman, Fell and Boekhout2011).
Results
A total of 20 isolates was obtained which were representing two genera of yeasts and one filamentous fungi belonging to two classes: Ascomycota (Articulospora sp., Thelebolus sp.) and Basidiomycota (Rhodotorula sp.).
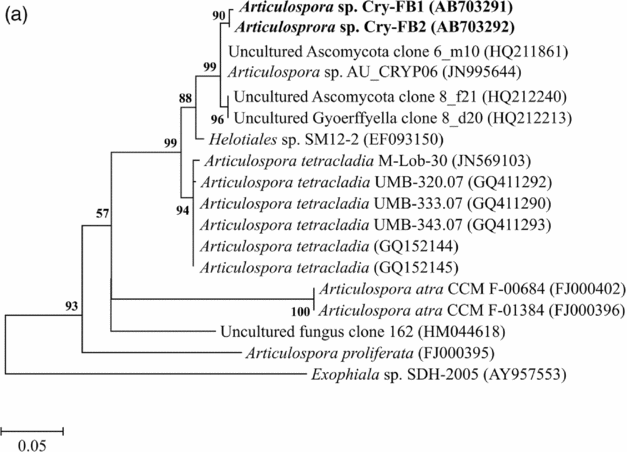
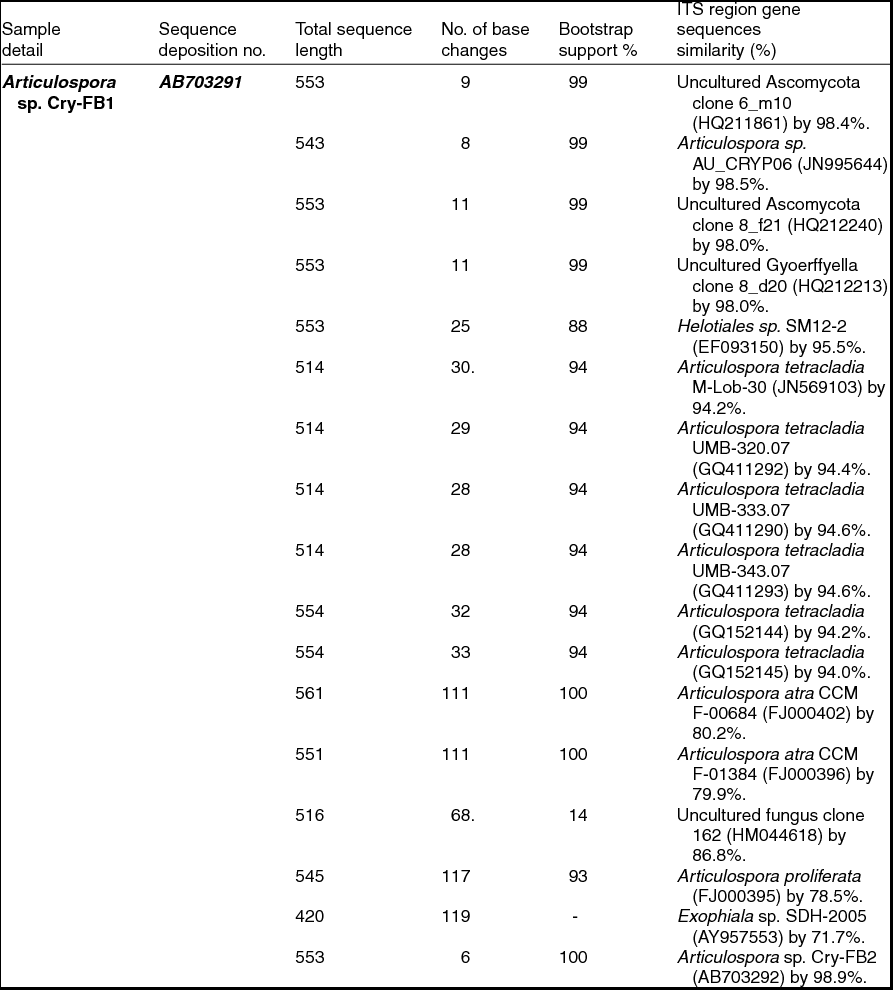
Sequencing of ITS region and subsequent BLAST search showed that isolates of Articulospora sp. Cry-FB1 and Cry-FB2 closely resembled Articulospora tetracladia M-Lob-30 (JN569103) by 94.2%., A. tetracladia UMB-320.07 (GQ411292) by 94.4%., A. tetracladia UMB-333.07 (GQ411290) by 94.6%, A. tetracladia UMB-343.07 (GQ411293) by 94.6%, A. tetracladia (GQvv152144) by 94.2%, A. tetracladia (GQ152145) by 94.0% and with Articulospora sp. AU_CRYP06 (JN995644) by 98.5% (Fig. 2a, Table 1a).

Fig. 2a. Phylogenetic analysis of Articulospora sp. using ITS region. The accession numbers of isolates are shown in parentheses. Tree was constructed with neighbor-joining method. The significance of each branch is indicated by a bootstrap value. The scale bar is estimated substitutions per nucleotide position.
Table 1a. Identification of Articulospora species (using ITS region), total sequence lengths after alignment, % sequence similarities, number of positions with base changes.

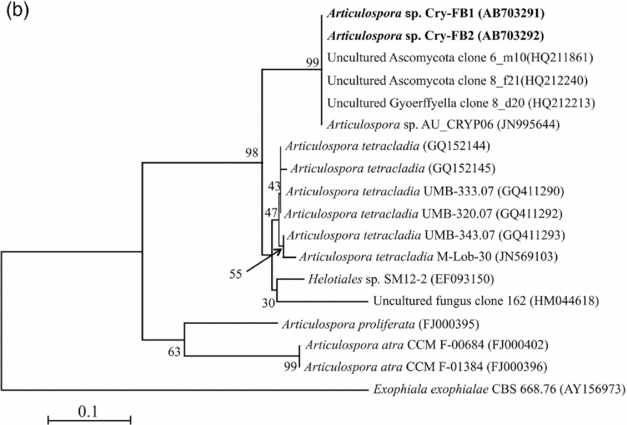
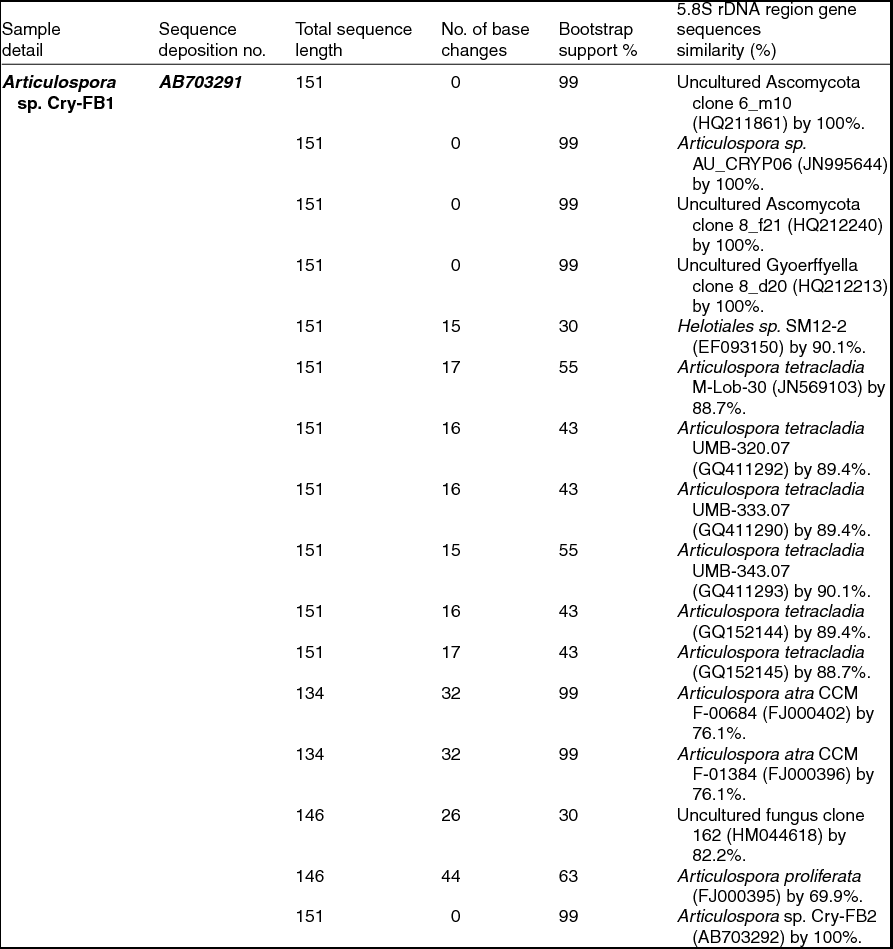
To confirm the novelty of Articulospora sp. Cry-FB1 and Cry-FB2 strains, the sequences of a more stable region (5.8S rRNA) which has slower evolutionary change rates were further analysed. 5.8S rRNA region showed closest sequences similarity (%) with Articulospora tetracladia M-Lob-30 (JN569103) by 88.7%, A. tetracladia UMB-320.07 (GQ411292) by 89.4%, A. tetracladia UMB-333.07 (GQ411290) by 89.4%, A. tetracladia UMB-343.07 (GQ411293) by 90.1%, A. tetracladia (GQ152144) by 89.4% and A. tetracladia (GQ152145) by 88.7% (Fig 2b, Table 1b). These analyses confirmed that Articulospora sp. Cry-FB1 (AB703291) and Cry-FB2 (AB703292) are novel strains. In phylogenetic tree the isolates Articulospora sp. Cry-FB1 and Cry-FB2 presented as novel species are yet to be established. The total sequence lengths after alignment, % sequence similarities, and number of positions with base changes are summarised in Tables 1a and 1b.

Fig. 2b. Phylogenetic analysis of Articulospora sp. using 5.8S rDNA region. The accession numbers of isolates are shown in parentheses. Tree was constructed with NJ method. The significance of each branch is indicated by a bootstrap value. The scale bar is estimated substitutions per nucleotide position.
Table 1b. Identification of Articulospora species (using 5.8S rDNA region), total sequence lengths after alignment, % sequence similarities, number of positions with base changes

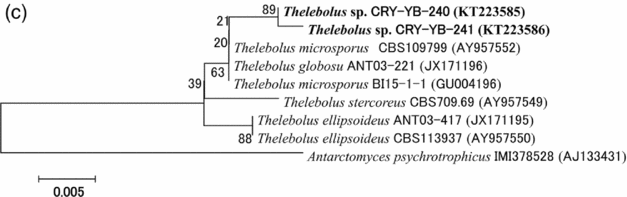
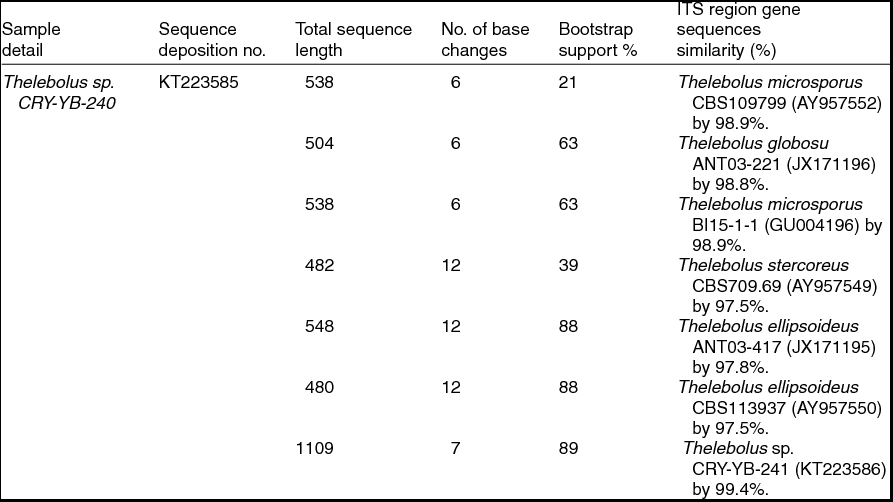
Sequence analyses of isolates Thelebolus sp. CRY-YB-240 (KT223585) and Thelebolus sp.CRY-YB-241 (KT223586) resembled Thelebolus microsporus CBS109799 (AY957552). Rhodotorula sp. Cry-FB3 indicated their closest relationship to the species of Rhodotorula svalbardensis Cry-YB-1 (AB734690). The phylogenetic tree of Thelebolus sp. is shown in Fig. 2c. The total sequence lengths after alignment, % sequence similarities, and number of positions with base changes of Thelebolus sp. CRY-YB-240, Thelebolus sp.CRY-YB-241 and closely related strains are shown in Table 2.

Fig. 2c. Phylogenetic tree of Thelebolus sp. using ITS region. The accession numbers of isolates are shown in parentheses. Tree was constructed with NJ method. The significance of each branch is indicated by a bootstrap value. The scale bar is estimated substitutions per nucleotide position.
Table 2. Identification of Thelebolus species (using ITS region), total sequence lengths after alignment, % sequence similarities, number of positions with base changes.

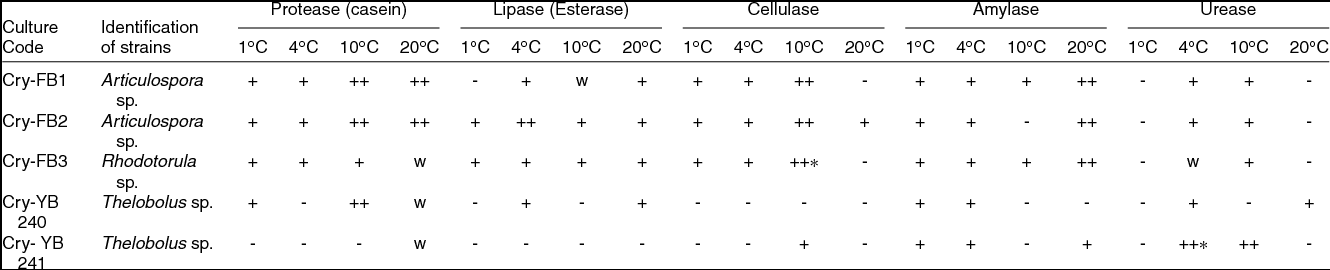
Most of the fungi isolated from the AB and VB glaciers cryoconites were capable of growing between 1 and 25˚C, indicating that fungi are cryophilic in nature. Isolates have shown salt tolerance ranging from 1–7% NaCl. It was worth investigating the potential of Articulospora sp, Rhodotorula sp. and Thelebolus sp. in terms enzyme production. The filamentous fungus like Articulospora sp., expressed high amylase, cellulase, lipase and protease activities whereas yeast Rhodotorula sp. showed high amylase and cellulase activity. Thelebolus sp. showed protease and urease activities (Table 3). The change in colour from orange to pink (Kurtzman and others Reference Kurtzman, Fell and Boekhout2011) was considered positive (Fig. 1h).
Table 3. Screening of fungal isolates for enzymes ability from Austre and Vestre Brøggerbreen glaciers of Svalbard.

++, Strong positive; +, Positive; W, weak; -, Negative.
[Halo zone size (1mm-15 mm) = +, (16-20 mm & above) = ++]
Discussion
Literature on culturable fungal diversity and biotechnological potential of isolates from glacier cryconites is scarce. Recently, Singh and Singh (Reference Singh and Singh2012) reported five genera (Cryptococcus, Mrakia, Rhodotorula., Phialophora and Articulospora) from ML glacier, while Edward and others (Reference Edwards, Douglas and Anesio2013) reported two genera (Articulospora and Varicosporium) from AB, ML and VB glaciers. In this study, fungi from three genera (Articulospora, Thelebolus, Rhodotorula) from AB and VB glaciers were isolated and their enzymatic potential analysed. The ecological role of these fungi in the cryoconite holes possibly drive the process of organic macromolecule degradation through enzyme secretion, thereby assisting in nutrient cycling in these supraglacial environments. Edward and others (Reference Edwards, Douglas and Anesio2013) mentioned the role of Articulospora and Varicosporium as decomposers in the carbon dynamics of cryoconite holes.
The capability of subsisting at the midst of cryoconite holes at low temperatures (0.1-1.5°C), and maintaining physiological processes depend on the bio-macromolecules like enzymes that are cold-active. Most of the catabolic and anabolic reactions are energy consuming process and possibly the enzymes produced by the fungal isolates (Articulospora, Thelebolus, Rhodotorula) help them in cold adaptation mechanism and survival in oligotrophic glacier environment. The role of psychrophilic enzymes in cold adaptation has been reported earlier (Feller Reference Feller and Gerday2003; Lonhienne and others Reference Lonhienne, Gerday and Feller2000). There are various interactions at the subcellular level, in which certain enzymes act as inducers for chains of further processes which are related to precursors of other reaction chains (Booth Reference Booth, Lengeler, Drews and Schlegel1999; Orange Reference Orange1994). During extreme environmental conditions, enzymes regulate the osmotic status of the cell by producing sugars, sugar alcohols, and polyols. Thus, enzyme producing ability of fungal isolates of cryoconites has an ecological significance.
The yeast colonies isolated from cryoconites showed orange and brown colour due to the presence of pigments which probably play some roles in maintaining the membrane fluidity. It is known that the modulation of membrane fluidity is brought about by changing the levels of polar and non-polar carotenoids (Jagannadham and others Reference Jagannadham, Chattopadhyay and Subbalakshmi2000). Thus, it seems that the pigments interacting with cell membranes increase its rigidity and enable cold-adaptation in cryoconite environment.
The screening results for various enzymes such as amylase, cellulase, lipase, protease, and urease of the isolates exhibited strong enzyme activities. These enzymes from AB and VB glaciers fungi promise biotechnological potentials. Studies on the cold-active enzymes from the polar regions are still fragmentary (Bej and Mojib Reference Bej, Mojib, Bej, Aislabie and Atlas2009; Männistö and Häggblom Reference Männistö and Häggblom2006; Medigue and others Reference Medigue, Krin and Pascal2005, Singh and others Reference Singh, Tsuji and Singh2013b) although they find applications in health, agriculture and industry (Feller and Gerday Reference Feller and Gerday2003). Further studies on purification and characterisation of enzymes of these fungal isolates (Articulospora, Thelebolus, Rhodotorula) would provide better understanding about the biotechnological potentials, and its application in health, agriculture and industry.
Based on DNA sequence analyses, Articulospora sp. Cry-FB1 (AB703291) and Articulospora sp. Cry-FB2 (AB703292) are potentially novel strains with little 5.8S rDNA sequence homology to known species. Characterisation of these putative novel species of Articulospora svalbardensis sp. nov. will be addressed in a separate study.
Acknowledgments
We would like to thank the Directors, BITS, NIPR and NCAOR for facilities. Authors are thankful to Department of Science and Technology (DST) India for providing the financial support [SR/WOSA/LS-419/2013(G)]. The authors also acknowledge Dr. S.M. Singh of Polar Biology Laboratory, National Centre for Antarctic Ocean & Research (NCAOR) Goa for his valuable suggestions.