Introduction
The physical properties of DNA are interrogated in virtually every process that involves storage or manipulation of the genetic information encoded in the double-helix. DNA packaging inside eukaryotic nuclei requires bending of the DNA around the histone octamer; and gene regulation in bacteria often involves DNA looping in regulatory regions. Transcription and DNA replication require local melting of the double-helix and produce a torsional stress in the DNA that is absorbed via the formation of plectonemes. In the pursue of a quantitative characterization of these and other biological processes, intensive research has been devoted to study the mechanical properties of DNA (Bloomfield et al., Reference Bloomfield, Crothers and Tinoco2000). In particular, in the last few decades, our understanding of DNA mechanics has been propelled by the advent of single-molecule techniques, which enable manipulating and/or measuring individual DNA molecules. Initial single-molecule experiments on DNA stretching and supercoiling (Smith et al., Reference Smith, Finzi and Bustamante1992; Strick et al., Reference Strick, Allemand, Bensimon, Bensimon and Croquette1996) showed that the mechanical properties of long (several kilobase pairs) DNA molecules are well described by polymer models that neglect the microscopic details of the duplex (Marko and Siggia, Reference Marko and Siggia1994, Reference Marko and Siggia1995a, Reference Marko and Siggia1995b). Since then, these polymer models have resulted extremely useful to quantify, for instance, the action of proteins on the DNA, and are currently employed to calibrate single-molecule biophysical instruments on a daily basis (Bouchiat et al., Reference Bouchiat, Wang, Allemand, Strick, Block and Croquette1999; Bustamante et al., Reference Bustamante, Bryant and Smith2003; Madariaga-Marcos et al., Reference Madariaga-Marcos, Hormeño, Pastrana, Fisher, Dillingham and Moreno-Herrero2018).
Despite the unquestionable success of polymer models, recent single-molecule assays are encouraging an alternative, i.e. microscopic description of DNA flexibility (Wiggins et al., Reference Wiggins, Heijden T, Moreno-Herrero, Spakowitz, Phillips, Widom, Dekker and Nelson2006; Lipfert et al., Reference Lipfert, Kerssemakers, Jager and Dekker2010; Vafabakhsh and Ha, Reference Vafabakhsh and Ha2012; Lebel et al., Reference Lebel, Basu, Oberstrass, Tretter and Bryant2014; Shon et al., Reference Shon, Rah and Yoon2019; Pyne et al., Reference Pyne, Noy, Main, Velasco-Berrelleza, Piperakis, Mitchenall, Cugliandolo, Beton, Stevenson, Hoogenboom, Bates, Maxwell and Harris2020). Ultimately, DNA is a highly sophisticated molecule with fine molecular features such as major and minor grooves, stacking interactions, or hydrogen bond donor and acceptors, that are inextricably linked to the flexibility of the double-helix. For example, dinucleotides that present weak stacking interactions are prone to induce a sharp bend in the DNA (Olson et al., Reference Olson, Gorin, Lu, Hock and Zhurkin1998); sequences with a narrow minor groove resist mechanical deformations (Nelson et al., (Reference Nelson, Finch, Luisi and Klug1987); and exotic hydrogen bonds found in certain sequences can assist complex deformations of the double-helix (Dans et al., Reference Dans, Faustino, Battistini, Zakrzewska, Lavery and Orozco2014; Pasi et al., Reference Pasi, Maddocks, Beveridge, Bishop, Case, Cheatham, Iii Dans, Jayaram, Lankas, Laughton, Mitchell, Osman, Orozco, Pérez, Petkevičiūtė, Spackova, Sponer, Zakrzewska and Lavery2014). Consequently, despite behaving as a homogeneous polymer at long scales, DNA presents an intricate flexibility at short distances that is strongly dependent on the nucleotide sequence (see Fig. 1). In addition, non-canonical DNA base pairs including e.g. methylated cytosines or mismatched base pairs, can substantially alter the mechanics of DNA at such short scales (see Fig. 1). Thus, given that DNA:protein interactions usually occur at the nanometer scale, it is paramount to deepen the molecular description of DNA flexibility, which will possibly provide additional insights into biological phenomena beyond the reach of classical long-range polymer approaches. Moreover, a molecular characterization of DNA mechanics may pave the way for expanding the nanotechnological applications of DNA as building material.
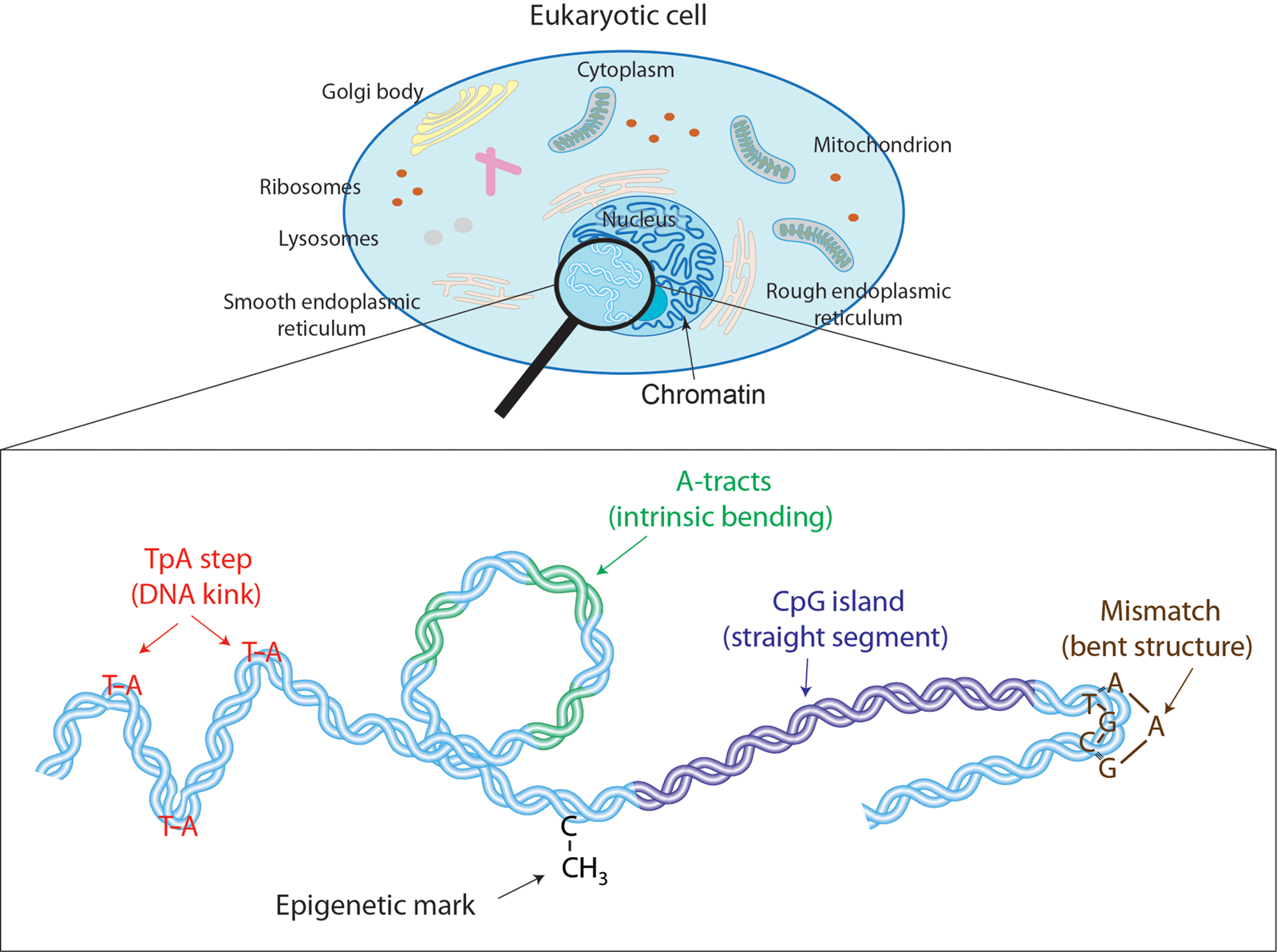
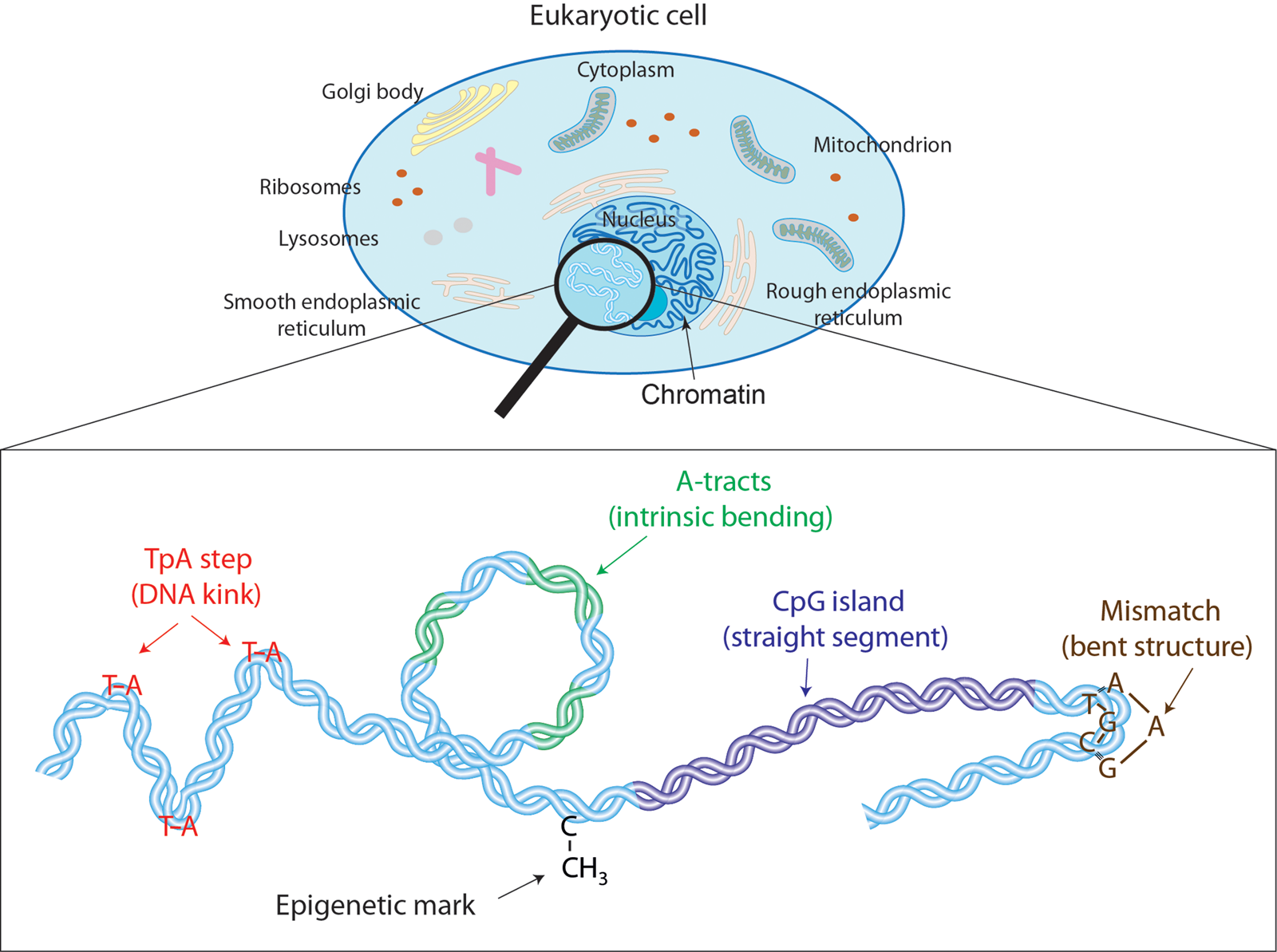
Fig. 1. Effectors of DNA mechanical properties at short-length scales. DNA is highly compacted in the chromatin inside the nucleus of a cell. We will show along the text that multiple effectors have been described to modulate the DNA mechanical properties at short-length scales. These examples comprise kinkable TpA steps, intrinsic bending by A-tracts, rigid CGIs, mismatches that produce strong bending, and cytosine methylation that exert a versatile role in DNA physical properties.
Here, we review recent studies that are advancing our understanding of single-molecule DNA mechanics from such a molecular perspective. We focus on two approaches that are driving major progress in this field. First, the development of new single-molecule assays that permit assessing novel mechanical properties of DNA, especially at short-length scales. Second, the synergistic combination of single-molecule experiments and computer simulations, most notably molecular dynamics (MD), that model the mechanics of DNA with atomic details. This review aims to discuss recent studies that illustrate the capability of these two approaches at providing novel relevant insights into double-stranded DNA mechanics. Thus, it is not our intention to deepen in any particular aspect of DNA mechanics or to include non-helical DNA structures, such as G-quadruplexes or i-motifs (see Abou Assi et al., Reference Abou Assi, Garavis, Gonzalez and Damha2018; Mandal et al., Reference Mandal, Hoque, Mao, Yang and Lin2019 for reviews on these topics).
This review has been divided into three sections. In Section ‘Sequence-dependent DNA mechanics’, we comment on how the nucleotide sequence affects the physical properties of DNA. We review sequences that facilitate or preclude DNA bending; sequences prone to adopt a double-helical structure that differs from the canonical B-DNA; and sequences where DNA melting is more favorable. In Section ‘Chemical modifications and DNA mechanics’, we discuss the effect of modifications of the canonical duplex, in particular, methylated cytosines and DNA mismatches. In Section ‘Mechanical properties of dsRNA: unexpected differences with dsDNA’, we cover recent studies that are revealing interesting differences between the mechanics of DNA and double-stranded RNA (dsRNA), the other double-helical carrier of genetic information that is found in nature. Finally, we present the conclusions of the review and suggest future lines of research.
The findings on DNA mechanics hereby reviewed have often provided a new perspective on diverse biological systems, as discussed throughout the text. An exciting challenge for the near future will be to exploit our ever-growing knowledge on nucleic acids mechanics for controlled design of functional DNA and RNA nanostructures.
Sequence-dependent DNA mechanics
In single-molecule experiments, the mechanical properties of DNA are commonly quantified by means of its persistence length, P DNA. If one thinks of a long shoelace or a (cooked) spaghetti immersed in a swimming pool, one does not picture a perfectly straight line, but rather, a somewhat coiled object. The same occurs for DNA. The size of the DNA polymer in solution, as given by the distance between its two ends, is quantified by P DNA. An alternative – perhaps more intuitive – way of looking at P DNA is the threshold DNA length above which the molecule will start to bend. In other words, DNA molecules shorter than P DNA are expected to be essentially straight. Several single-molecule experimental setups, including magnetic tweezers (MT), optical tweezers (OT) (Smith et al., Reference Smith, Finzi and Bustamante1992, Reference Smith, Cui and Bustamante1996; Bustamante et al., Reference Bustamante, Marko, Siggia and Smith1994; Wang et al., Reference Wang, Yin, Landick, Gelles and Block1997), and atomic force microscopy (AFM) imaging (Rivetti et al., Reference Rivetti, Guthold and Bustamante1996; Heenan and Perkins, Reference Heenan and Perkins2019), allow extracting the value of P DNA with high accuracy. These and many other single-molecule techniques yield, under standard buffer conditions, a consensual value of P DNA~50 nm, which corresponds to ~150 bp (e.g. the length of DNA in the nucleosome core).
Note, however, that the accepted value of P DNA~150 bp contrasts with several biological evidences where the DNA is severely bent at distances comparable to or shorter than 150 bp. Examples of such bends include DNA loops at regulatory regions or the wrapping of nucleosomal DNA around the histone core (reviewed in Garcia et al., Reference Garcia, Grayson, Han, Inamdar, Kondev, Nelson, Phillips, Widom and Wiggins2007). According to the commonly accepted worm-like chain (WLC) model, such short-scale deformations would be prohibitive for a DNA molecule with P ~150 bp.
This conundrum motivated studies on the local mechanics of single DNA molecules (Wiggins et al., Reference Wiggins, Heijden T, Moreno-Herrero, Spakowitz, Phillips, Widom, Dekker and Nelson2006; Vafabakhsh and Ha, Reference Vafabakhsh and Ha2012). Such studies revealed a striking ability of the duplex to adopt strongly bent conformations, an aspect that was not contemplated by the standard WLC model (Wiggins et al., Reference Wiggins, Heijden T, Moreno-Herrero, Spakowitz, Phillips, Widom, Dekker and Nelson2006; Vafabakhsh and Ha, Reference Vafabakhsh and Ha2012). Although the quantitative aspects of short-scale DNA bending remain a subject of debate, it is clear that sharp DNA bending can proceed via concerted localized distortions of the duplex, such as DNA kinks (Vologodskii and Frank-Kamenetskii, Reference Vologodskii and Frank-Kamenetskii2013). More interestingly, the nucleotide sequence strongly affects these local deformations (Vafabakhsh and Ha, Reference Vafabakhsh and Ha2012), in contrast to the long-range persistence length, which shows little sequence-dependent variation (Geggier and Vologodskii, Reference Geggier and Vologodskii2010). Namely, sequence effects on DNA mechanics appear amplified at short-length scales.
TpA dinucleotides are highly flexible
DNA kinks were predicted long ago (Crick and Klug, Reference Crick and Klug1975) and have been observed in crystal structures of a number of DNA:protein complexes (Berman et al., Reference Berman, Olson, Beveridge, Westbrook, Gelbin, Demeny, Hsieh, AR and B1992; Olson et al., Reference Olson and Zhurkin1998), including nucleosomal DNA (Olson and Zhurkin, Reference Olson and Zhurkin2011); and also in several MD simulations (Lankaš et al., Reference Lankaš, Lavery and Maddocks2006; Curuksu et al., Reference Curuksu, Zacharias, Lavery and Zakrzewska2009; Irobalieva et al., Reference Irobalieva, Fogg, Catanese, Sutthibutpong, Chen, Barker, Ludtke, Harris, Schmid, Chiu and Zechiedrich2015; Harrison et al., Reference Harrison, Romano, Ouldridge, Louis and Doye2019). The biological consequences of DNA kinks are diverse, and have been reviewed in the context of DNA wrapping in the nucleosome core (Richmond and Davey, Reference Richmond and Davey2003; Olson and Zhurkin, Reference Olson and Zhurkin2011) and of specific DNA:protein interactions (Rohs et al., Reference Rohs, Jin, West, Joshi, Honig and Mann2010). Crystal structures of DNA:protein complexes reveal that kinks are more favorable in pyrimidine-purine steps, CpG and CpA, and particularly in the TpA step, which has shown larger flexibility (Olson et al., Reference Olson, Gorin, Lu, Hock and Zhurkin1998) (see Table 1 for nomenclature). However, structural studies provide a static description of the DNA and thus, are of limited use in order to address the dynamics of kink formation.
Table 1. Nomenclature used for DNA and dsRNA sequences
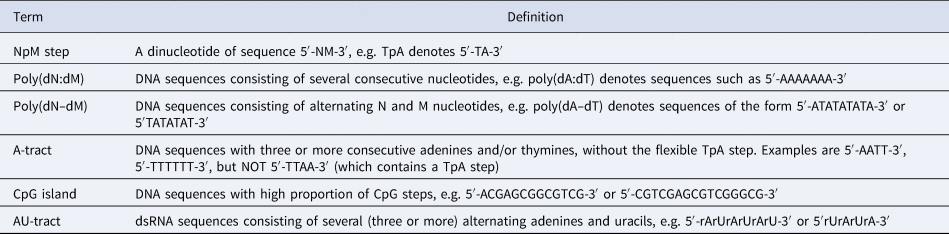
Note: The sequences shown here refer to a canonical Watson–Crick double-helix where the reverse strand is omitted for clarity, e.g. the sequence 5′-AGTACCC-3′ refers to a DNA double-helix with forward strand 5′-AGTACCC-3′ and reverse strand 5′-GGGTACT-3′. Ribonucleotides are referred to as rA, rC, rU, and rG.
The development of novel single-molecule assays to interrogate the flexibility of short DNA molecules is a promising tool to probe the impact of kinkable pyrimidine-purine steps on the physical properties of DNA. Of particular relevance is the creative single-molecule assay developed by Vafabakhsh and Ha (Reference Vafabakhsh and Ha2012). By attaching a pair of fluorescence resonance energy transfer (FRET) dyes to DNA molecules and measuring the FRET signal, the authors were able to observe single-molecule DNA cyclization in real time (see Fig. 2a). In that study, the authors showed that the cyclization dynamics, as quantified by the J-factor, was much faster than the WLC prediction, supporting the existence of strongly bent DNA structures, as proposed in a previous AFM study (Wiggins et al., Reference Wiggins, Heijden T, Moreno-Herrero, Spakowitz, Phillips, Widom, Dekker and Nelson2006).
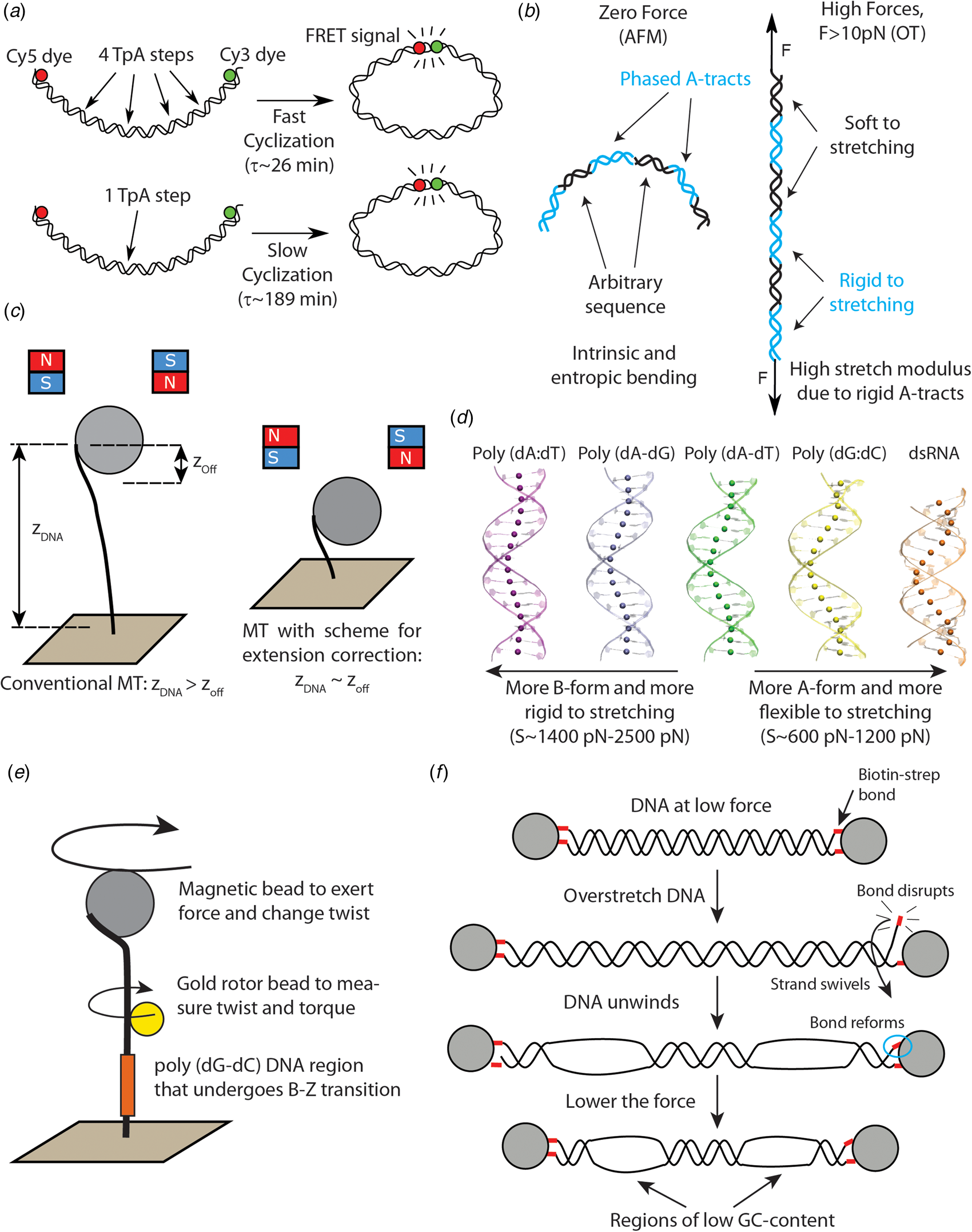
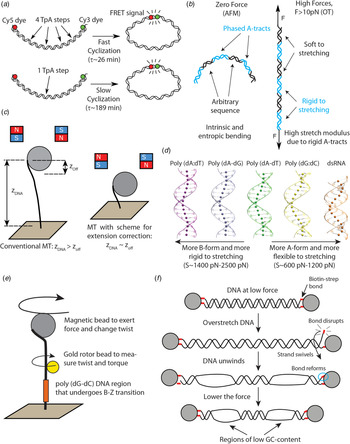
Fig. 2. Recent single-molecule and MD studies on sequence-dependent DNA mechanics. (a) The single-molecule assay developed by Vafabakhsh and Ha (Reference Vafabakhsh and Ha2012) consists of a pair or FRET dyes attached to the extremes of a DNA molecule with cohesive ends. Cyclization results in an increase in the FRET signal, thus enabling measuring single-molecule cyclization in real time. TpA steps were found to increase DNA flexibility, resulting in faster cyclization kinetics Ngo et al. (Reference Ngo, Zhang, Zhou, Jaya G and Ha2015). (b) Cartoon illustrating the complex mechanical properties of A-tract sequences. A-tracts located in phase with the DNA helical pitch induce a global macroscopic curvature in the molecule. When the molecule is subjected to high forces, the bends are straightened and the A-tracts are found to present a large stretching rigidity. Thus, A-tracts induce DNA bending, but the tracts themselves are rigid at a local level. Adapted from Marin-Gonzalez et al. (Reference Marin-Gonzalez, Pastrana, Bocanegra, Martín-González, Vilhena, Pérez, Ibarra, Aicart-Ramos and F2020a). (c) MT are usually limited to measurements of DNA molecules with contour length longer than the bead radius. This limitation is overcome in the MT scheme for extension correction developed in Shon et al. (Reference Shon, Rah and Yoon2019), which enables estimating the anchor point of the DNA on the bead. Using this correction scheme, accurate MT force–extension curves can be obtained for DNA molecules as short as ~200 bp. (d) The crookedness curvature reported in Marin-Gonzalez et al. (Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a) is responsible for sequence-dependent variations of the DNA extension and distinguishes between A- and B-DNA conformations. Curved sequences, such as poly(dG:dC) are A-like and flexible to stretching; straight sequences, such as poly(dA:dT) are highly B-like and rigid to stretching. Adapted from Marin-Gonzalez et al. (Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a). (e) The gold rotor bead assay from Lebel et al. (Reference Lebel, Basu, Oberstrass, Tretter and Bryant2014) combines high-resolution torque spectroscopy and controlled stretching and supercoiling of DNA molecules. A magnetic bead permits exerting force and supercoiling DNA molecules; whereas a gold bead attached to the side of the molecule reports on its twist and torque. This system has been used to study the B–Z transition of a poly(dG–dC) sequence with high temporal resolution. (f) Assay for DNA supercoiling using OT, as reported in King et al. (Reference King, Burla, Peterman and Wuite2019). Upon overstretching, one of the biotin–streptavidin interactions that attach the DNA to the optical beads is disrupted. The DNA unwinds and then the bond reforms, trapping the DNA in a negatively supercoiled state. When the force is lowered, melting bubbles are observed in regions of low GC-content.
More importantly, this single-molecule cyclization assay was employed to explore the effect of the nucleotide sequence on DNA flexibility. In a later study, the same group studied the cyclization dynamics of the two halves of the 601 Widom sequence (a strong nucleosome positioning sequence) (Ngo et al., Reference Ngo, Zhang, Zhou, Jaya G and Ha2015). They found that the left half, which contains four kinkable TpA steps, was highly flexible, whereas the right half, which only contains one of such steps, was relatively rigid (see Fig. 2a). Notably, when this rigid half was modified to include three additional TpA steps, it became much more flexible, albeit it was still more rigid than the left, flexible half. This experiment shows that even few TpA steps can substantially impact the flexibility of DNA molecules comprising tens of base pairs. Further single-molecule studies should aim to address whether this effect is also present in CpG and CpA steps, which have also been traditionally identified as highly flexible (Olson et al., Reference Olson, Gorin, Lu, Hock and Zhurkin1998).
A deeper understanding of the physical properties of DNA sometimes offers a fresh perspective on certain biological processes. This is well illustrated in the study by Ngo et al. (Reference Ngo, Zhang, Zhou, Jaya G and Ha2015). Besides the aforementioned cyclization experiments, the authors used a combined setup of OT and single-molecule FRET (smFRET) to observe single unwrapping events in nucleosomes. This novel assay permits visualizing which side of the nucleosomal DNA is detached from the histone core when tension is applied. They observed that reconstituted nucleosomes with the aforementioned 601 sequence showed highly asymmetric unwrapping. In particular, upon the action of an external force, the right rigid half of the nucleosomal DNA was significantly more prone to unwrap than the left, flexible half. On the contrary, nucleosomes reconstituted with the 601 sequence containing additional TpA steps in the right half showed an approximate symmetric unwrapping. Namely, when the two halves of nucleosomal DNA had a similar flexibility, nucleosome unwrapping occurred stochastically from either side. Since the publication of the study by Ngo et al. (Reference Ngo, Zhang, Zhou, Jaya G and Ha2015), a number of experimental and computational studies have addressed the link between DNA flexibility and transient nucleosome unwrapping, or nucleosome breathing (Lequieu et al., Reference Lequieu, Córdoba, Schwartz and De Pablo2016; Zhang et al., Reference Zhang, Zheng, Papoian and Wolynes2016; Chen et al., Reference Chen, Tokuda, Topping, Meisburger, Pabit, Gloss and Pollack2017; Culkin et al., Reference Culkin, De Bruin, Tompitak, Phillips and Schiessel2017; Mauney et al., Reference Mauney, Tokuda, Gloss, Gonzalez and Pollack2018; Winogradoff and Aksimentiev, Reference Winogradoff and Aksimentiev2019). These studies have revealed that, besides its well-known role in positioning nucleosomes (Widom, Reference Widom2001; Segal et al., Reference Segal, Fondufe-Mittendorf, Chen, Thåström, Field, Moore, Wang and Widom2006; Kaplan et al., Reference Kaplan, Moore, Fondufe-Mittendorf, Gossett, Tillo, Field, Leproust, Hughes, Lieb, Widom and Segal2009), the DNA sequence can largely affect the accessibility of certain regions within nucleosomal DNA via changes in the flexibility of the duplex.
A dual role of A-tracts in DNA flexibility
In contrast to the flexibility of TpA steps, other DNA sequence motifs are thought to be extremely rigid. The most well-known examples are A-tracts, a kind of DNA sequence that consists of three or more adenines and thymines without the flexible TpA step. Poly(dA:dT)s, a particular case of A-tracts, possess a strong nucleosome depleting character that has been associated to a presumably high rigidity of this motif, as predicted from structural studies and atomistic simulations (Nelson et al., Reference Nelson, Finch, Luisi and Klug1987; Haran and Mohanty, Reference Haran and Mohanty2009; Segal and Widom, Reference Segal and Widom2009; Dršata et al., Reference Dršata, Špačková, Jurečka, Zgarbová, Šponer and Lankaš2014).
The flexibility of A-tracts in general, and poly(dA:dT)s in particular, are controversial, because they are inextricably intertwined with the long-known intrinsic bending induced by these sequences (see, e.g. Zhang and Crothers, Reference Zhang and Crothers2003; Thompson and Travers, Reference Thompson and Travers2004; Haran and Mohanty, Reference Haran and Mohanty2009; Peters and Maher, Reference Peters and Maher2010; Ortiz and de Pablo, Reference Ortiz and de Pablo2011). For example, previous AFM and tether particle motion experiments indicated that A-tracts have no enhanced bending rigidity, showing a standard persistence length of ~50 nm (Rivetti et al., Reference Rivetti, Walker and Bustamante1998; Brunet et al., Reference Brunet, Chevalier, Destainville, Manghi, Rousseau, Salhi, Salome and Tardin2015a). However, these results contrast with coarse-grain simulations and single-molecule cyclization experiments, which suggest that A-tracts are rigid to bending (Vafabakhsh and Ha, Reference Vafabakhsh and Ha2012; Mitchell et al., Reference Mitchell, Glowacki, Grandchamp, Manning and Maddocks2017).
In a recent study, these paradoxical mechanical properties of A-tracts were comprehensively studied at the single-molecule level using a wealth of techniques: MT, OT, and AFM imaging in air and in liquid (Marin-Gonzalez et al., Reference Marin-Gonzalez, Pastrana, Bocanegra, Martín-González, Vilhena, Pérez, Ibarra, Aicart-Ramos and F2020a). The authors considered a sequence from the Caenorhabditis elegans genome with several repetitions of phased A-tracts, expected to display a strongly bent character (Fire et al., Reference Fire, Alcazar and Tan2006). Indeed, AFM measurements demonstrated that phased A-tracts induce an intrinsic bend in the DNA that could be well-described with a variant of the WLC model that includes intrinsic bending (see Fig. 2b) (Rezaei et al., Reference Rezaei, Lyons and Forde2018; Marin-Gonzalez et al., Reference Marin-Gonzalez, Pastrana, Bocanegra, Martín-González, Vilhena, Pérez, Ibarra, Aicart-Ramos and F2020a). Moreover, both AFM and OT measurements independently showed that, at long-length scales, the A-tract molecule was not particularly rigid (nor flexible) to bending, showing a persistence length of 54 nm. However, as the A-tracts were subjected to high forces (F > 10 pN), an extraordinary rigidity started to emerge, that was quantified by a large stretch modulus (see Fig. 2b). Such high forces are expected to align the intrinsic A-tracts bends, thus enabling to probe the local rigidity of the A-tract structure. It was thus proposed that, although A-tract might not seem rigid at long distances, the stiff local structure of the A-tract would resist short-scale mechanical deformations, which are likely more determinant for nucleosome formation.
We can thus conclude that A-tracts play a dual role in DNA flexibility. On the one hand, they induce a static bending in the double-helix, but on the other hand the structure of the A-tract itself appears rigid. It is tempting to state that the balance of these two effects can be regulated by the distribution of A-tracts along a given DNA region. That is, several short A-tracts in phase with the helical pitch would amplify the bending, whereas a long (i.e. >20 bp) individual A-tract would stiffen the DNA. This intriguing dual mechanism appears to be exploited in vivo in the context of nucleosome positioning: short, phased A-tracts are enriched in nucleosome positioning sequences Rohs et al. (Reference Rohs, West, Sosinsky, Liu, Mann and Honig2009), whereas long poly(dA:dT)s are characteristic of nucleosome depleting regions (Segal and Widom, Reference Segal and Widom2009).
CpG islands are rigid DNA regions depleted of nucleosomes
Another example of DNA motif whose biological functions might depend on peculiar mechanical properties are CpG islands (CGI), GC-rich DNA regulatory regions with a high proportion of CpG steps. In vertebrates, CGI are ubiquitous sites of transcription initiation, a feature that has been linked with a deficient assembly of CGI into nucleosomes (both in vivo and in vitro) (Ramirez-Carrozzi et al., Reference Ramirez-Carrozzi, Braas, Bhatt, Cheng, Hong, Doty, Black, Hoffmann, Carey and Smale2009; Deaton and Bird, Reference Deaton and Bird2011). As for poly(dA:dT), it is likely that the depletion of nucleosomes at CGI is related to a peculiar rigidity of these sequences (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a; Shon et al., Reference Shon, Rah and Yoon2019; Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017).
Recent developments in single-molecule techniques have enhanced our understanding of the mechanical properties of CGI. Shon et al. (Reference Shon, Rah and Yoon2019) developed a novel scheme for in-situ correction of the DNA extension in MT, which enables obtaining accurate force–extension curves of DNA molecules as short as 198 bp. This development extends the capability of conventional MT, which are usually limited to DNA molecules longer than ~1 kbp; and thus optimizes this technique for better exploration of sequence effects on DNA flexibility (see Fig. 2c). Using their novel correction scheme, Shon et al. found that a CGI DNA molecule possessed a large persistence length when compared with a control DNA of arbitrary sequence, indicative of a high bending rigidity of CGI. Interestingly, this and other studies have proposed that the flexibility of CGI is highly sensitive to the methylation state of these sequences (Marin-Gonzalez, Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a; Shon et al., Reference Shon, Rah and Yoon2019; Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017), suggesting that epigenetic marks might exert regulatory functions in CGI via changes in the mechanical properties of the DNA. This will be discussed in Section ‘The impact of cytosine methylation on DNA physical properties’.
Despite these insightful studies, further inquiry is clearly needed to better understand the mechanical properties of CGI. First, there is some discrepancy among different single-molecule techniques regarding the large rigidity of CGI (Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017), which should be solved in future studies. Such studies should take into account that CGI not only have a high GC-content, but also a high density of CpG steps. In fact, it is expected that the frequency and distribution of CpG steps would play a more relevant role in the mechanics of DNA than the GC-content itself (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a). In parallel to single-molecule studies, computer simulations should aim at explaining the molecular mechanisms behind the proposed rigidity of CGI. This is a challenging task, given the complex deformability exhibited by the CpG step, which is highly anharmonic and dependent of its sequence context (Dans et al., Reference Dans, Faustino, Battistini, Zakrzewska, Lavery and Orozco2014).
Poly(dA–dT) and poly(dG:dC) adopt A-like DNA conformations
In addition to DNA flexibility, the nucleotide sequence can strongly impact the conformation of the double-helix. In particular, some sequences, such as poly(dG:dC), have been identified to possess structural features close to the A-form (A-philic), whereas others such as poly(dA:dT) are often classified as highly B-like (B-philic) (Lu et al., Reference Lu, Shakked and Olson2000). Yet, another class include poly(dA–dT) sequences, which adopt a so-called TA-DNA structure, or hybrid between B- and A-forms (Lu et al., Reference Lu, Shakked and Olson2000; Kulkarni and Mukherjee, Reference Kulkarni and Mukherjee2017). This rich conformational landscape of the double-helix is crucial for achieving DNA sequence recognition by several proteins that read specific features of the B- or A-forms (Lu et al., Reference Lu, Shakked and Olson2000; Rohs et al., Reference Rohs, Jin, West, Joshi, Honig and Mann2010) (see Table 2).
Table 2. Sequence-dependent DNA physical properties
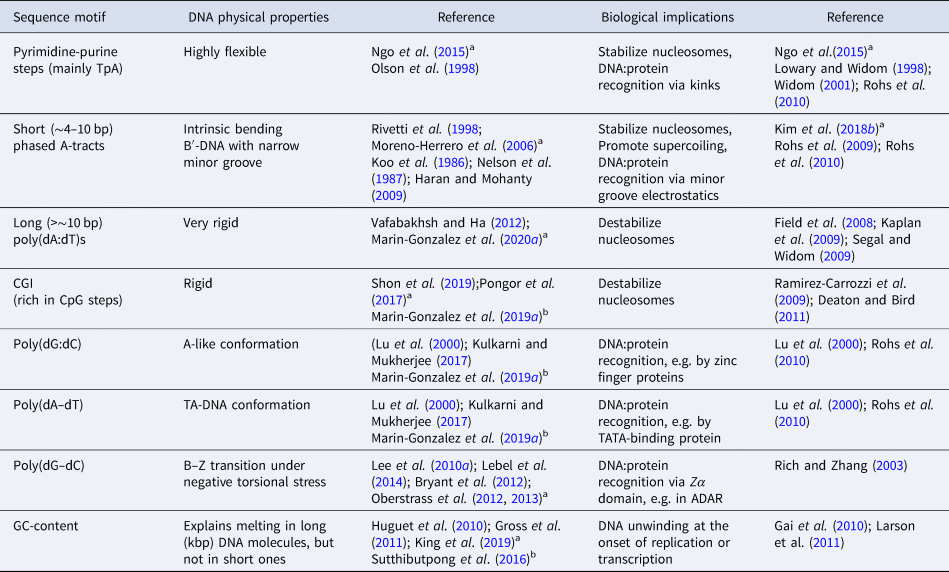
a Single-molecule experiments.
b Computer simulations.
Several structural parameters have been proposed as a measure of the A-philicity of a given DNA sequence, i.e. of its propensity to assume an A-like conformation (Lu et al., Reference Lu, Shakked and Olson2000; Waters et al., Reference Waters, Lu, Galindo-Murillo, Gumbart, Kim, Cheatham and Harvey2016; Kulkarni and Mukherjee, Reference Kulkarni and Mukherjee2017). However, calculating these parameters requires knowledge of the atomic structure of the molecule, which is often inaccessible to single-molecule experiments. Is it possible to assess the A-philicity of a DNA sequence without knowing its atomic structure?
Recently, a promising approach has been proposed. Inspired by previous studies on dsRNA (Chou et al., Reference Chou, Lipfert and Das2014; Lipfert et al., Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014), Marin-Gonzalez et al. (Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a) suggested a connection between the A-philicity of a DNA sequence and its stretching flexibility. The authors performed MD simulations of several DNA sequences and they observed significant variations in the extension from one sequence to another. They noticed that these changes in the extension were a consequence of a sequence-dependent DNA curvature, denoted crookedness, that reasonably correlated with the A-philicity of the sequence (see Fig. 2d). For example, the poly(dG:dC), which is highly A-philic, showed a short extension and large crookedness curvature; whereas the highly B-philic poly(dA:dT) was essentially straight. Interestingly, those DNA molecules that were more crooked, and more A-like, were more flexible to a stretching force; and vice versa, highly B-like sequences were more rigid. Among the latter are poly(dA:dT) and poly(dC–dG), whose large stretching rigidity are supported by recent experiments on A-tracts and CGI (Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017; Marin-Gonzalez et al., Reference Marin-Gonzalez, Pastrana, Bocanegra, Martín-González, Vilhena, Pérez, Ibarra, Aicart-Ramos and F2020a).
It is thus tempting to state that the stretching flexibility of a given DNA sequence is a good indicator of its A-philicity. This hypothesis is supported by naïve intuition. Note that a perfectly straight textbook B-DNA helix should be extremely hard to stretch. In such an ideal B-DNA, elongations beyond the contour length should result in unwinding and unstacking of the base pairs, which would easily disrupt the double-helix. This was already realized by Bustamante and coworkers in their seminal single-molecule MT experiment, where they proposed that the stretching elasticity of DNA should be a consequence of a ‘local curvature of the DNA axis’ (Smith et al., Reference Smith, Finzi and Bustamante1992). It is plausible that such ‘local curvature’ consists of sequence-dependent deviations from the straight B-form into a more curved A-form (see Fig. 2d).
However, further research is needed to better characterize the DNA A-philicity, crookedness and stretching flexibility, in order to establish a more solid connection between these DNA features. Along this line, a recent study by Lankas and coworkers has expanded our understanding on DNA crookedness, by assessing how this parameter depends on the temperature of the system (Dohnalová et al., Reference Dohnalová, Dršata, Šponer, Zacharias, Lipfert and Lankaš2020).
Poly(dG–dC) are hotspots of Z-DNA formation
Besides the right-handed helical structures discussed above, DNA is able to assume, under certain conditions, a left-handed conformation with a zig-zag backbone known as Z-DNA (Rich and Zhang, Reference Rich and Zhang2003). This DNA structure has been observed at sequences of alternating purines and pyrimidines, most notably poly(dG–dC), and is thought to require high salt concentrations or negative torsional stress in order to form (Rich and Zhang, Reference Rich and Zhang2003). The structural features of Z-DNA (and also Z-RNA) are recognized in vitro by certain proteins (those containing the so-called Zα domain, see Table 2). However, the in vivo relevance of Z-DNA is debated (Rich and Zhang, Reference Rich and Zhang2003; Herbert, Reference Herbert2019; Jiao et al., Reference Jiao, Wachsmuth, Kumari, Schwarzer, Lin, Eren, Fisher, Lane, Young, Kassiotis, Kaiser and Pasparakis2020). In this regard, some studies point toward transient formation of Z-DNA as regulatory mechanism during transcription, where the negative supercoils generated by RNA polymerase would stabilize this left-handed structure (Rich and Zhang, Reference Rich and Zhang2003).
A number of single-molecule studies have provided a quantitative description of the thermodynamics of Z-DNA formation (Lee et al., Reference Lee, Kim, Kim and Hong2010a; Lebel et al., Reference Lebel, Basu, Oberstrass, Tretter and Bryant2014; Bryant et al., Reference Bryant, Oberstrass and Basu2012; Oberstrass et al., Reference Bryant, Oberstrass and Basu2012, Reference Oberstrass, Fernandes, Lebel and Bryant2013). In a pioneer study, Lee et al. (Reference Lee, Kim, Kim and Hong2010a) combined MT and smFRET to simultaneously induce and measure B–Z transitions in a DNA molecule containing a (dG–dC)11 sequence, prone to assume a Z-form. These single-molecule experiments indicated that, in the presence of tension, Z-DNA formation requires only a small torsional strain, much lower than predictions based on bulk experiments. The study by Lee et al. thus suggested that Z-DNA might occur in vivo more often than expected, given that tension is likely to accumulate on DNA, e.g. during transcription.
The dynamics of the B- to Z-DNA transition have also been studied using the rotor bead tracking assay (Oberstrass et al., Reference Oberstrass, Fernandes and Bryant2012, Reference Oberstrass, Fernandes, Lebel and Bryant2013; Lebel et al., Reference Lebel, Basu, Oberstrass, Tretter and Bryant2014; Lipfert et al., Reference Lipfert, Van Oene, Lee, Pedaci and Dekker2015). In this experimental setup an extreme of the DNA is bound to a magnetic bead that permits stretching and supercoiling the duplex; and a second bead is attached to the side of the DNA at an intermediate position. The position and fluctuations of the latter bead inform, respectively, on the twist and torque of the system (Bryant et al., Reference Bryant, Stone, Gore, Smith, Cozzarelli and Bustamante2003; Gore et al., Reference Gore, Bryant, Nöllmann, Le, Cozzarelli and Bustamante2006a, Reference Gore, Bryant, Stone, Nöllmann, Cozzarelli and Bustamante2006b) (see Fig. 2e). Using different versions of this rotor bead assay, Oberstrass et al. obtained torque-twist measurements on DNA molecules with 22 and 50 bp-long GC repeats and observed transitions consistent with the formation of a Z-DNA structure at the poly(dG–dC) regions (Oberstrass et al., Reference Bryant, Oberstrass and Basu2012, Reference Oberstrass, Fernandes, Lebel and Bryant2013). These measurements were able to recapitulate relevant features of the B–Z transition, such as its high cooperativity. Recently, rotor bead assays using a gold probe have allowed a more detailed characterization of this phenomenon, unveiling the B- to Z-DNA transition with unprecedented temporal resolution (Lebel et al., Reference Lebel, Basu, Oberstrass, Tretter and Bryant2014).
Altogether, single-molecule assays have provided a rather comprehensive description of the B- to Z-DNA transition at GC repeats under torsional stress. Future single-molecule studies should aim to explore the dynamics of this transition at other repeating sequences, and the impact of Z-DNA binding proteins on this process. Recent experimental studies are starting to provide interesting insights into these questions (Kim et al., Reference Kim, Ganji, Kim, Van Der Torre, Abbondanzieri and Dekker2018a).
In addition to single-molecule experiments, MD simulations have also explored the B- to Z-DNA transition, shedding light on the atomistic mechanisms behind it (Lee et al., Reference Lee, Kim and Hong2010b; Moradi et al., Reference Moradi, Babin, Roland and Sagui2013; Chakraborty and Wales, Reference Chakraborty and Wales2017). Of particular relevance is the study by Moradi et al. (Reference Moradi, Babin, Roland and Sagui2013), which suggested that the B- to Z-DNA transition can proceed by means of several different mechanisms, rather than via a single pathway. In that study, MD simulations revealed a rich diversity of non-canonical DNA structures – such as an overstretched-like S-DNA, extensive base flipping or unpeeling of the two DNA strands – that act as intermediate states in the B- to Z-DNA transition. Future computational efforts should further elucidate the molecular aspects of this enigmatic process. Recent refinements of DNA force-fields which focused on improving the description of the Z-DNA structure (Zgarbová et al., Reference Zgarbová, Šponer, Otyepka, Cheatham, Galindo-Murillo and Jurečka2015) will offer an invaluable tool toward achieving this goal.
GC-content not always explains DNA melting
Negative torsional stress regulates access to the genetic information. DNA unwinding promotes disruption of the base pairing interactions, or DNA melting, resulting in larger exposure of the nucleobases. DNA is thus unwound by specialized enzymes that locally denature the double-helix in order to read its nucleotide sequence, e.g. in transcription (Larson et al., Reference Larson, Landick and Block2011), replication (Gai et al., Reference Gai, Chang and Chen2010), or CRISPR-mediated bacterial immunity (Szczelkun et al., Reference Szczelkun, Tikhomirova, Sinkunas, Gasiunas, Karvelis, Pschera, Siksnys and Seidel2014). In addition, DNA melting often occurs as a consequence of in vivo torsional strains, and can result in the formation of particular structures such as kinks or bubbles that are specifically recognized by proteins (Fogg et al., Reference Fogg, Randall, Pettitt, Sumners, Harris and Zechiedrich2012).
At a macroscopic level (several kbp), DNA melting can be well explained from the GC-content of a given sequence (Marmur and Doty, Reference Marmur and Doty1962; Vologodskii and Frank-Kamenetskii, Reference Vologodskii and Frank-Kamenetskii2018). Namely, higher GC-content sequences better resist melting due to the three hydrogen bonds of the G:C base pair compared to the two bonds of the A:T base pair. This idea is supported by a recent single-molecule study that employs a novel creative assay to generate negative supercoils on DNA using a conventional OT set-up (King et al., Reference King, Burla, Peterman and Wuite2019) (see Fig. 2f). In typical OT experiments, the DNA molecules are attached to the optically trapped beads by means of biotin–streptavidin bonds. King et al. found that, when torsionally constrained molecules are overstretched (at forces of ~110 pN), one of the biotin–streptavidin interactions can temporarily disrupt. When this occurs, the torsional stress accumulated on the DNA during overstretching can be partially relieved, by swiveling one DNA strand around the other (see Fig. 2f). Eventually, the broken biotin–streptavidin bond forms again, locking the DNA molecule in a negatively supercoiled state. The authors exploited this discovery to controllably unwind the DNA and induce melting events that were then detected using fluorescently labeled RPA protein (which strongly binds single-stranded DNA) as a reporter. Their results indicate that the sites of DNA melting are reasonably correlated with DNA regions of low GC-content. This finding is in agreement with experiments on DNA overstretching, where the thermodynamics of sequence-dependent DNA unpeeling could be predicted on the basis of the GC-content (Gross et al., Reference Gross, Laurens, Oddershede, Bockelmann, Peterman and Wuite2011).
At the microscopic level (few base pairs), however, torsion-induced DNA melting remains poorly understood. For example, studies on single-molecule unzipping of DNA hairpins suggest that, in such short DNA molecules, GC-content might not be a determinant factor of DNA melting (Huguet et al., Reference Huguet, Bizarro, Forns, Smith, Bustamante and Ritort2010; Camunas-Soler et al., Reference Camunas-Soler, Ribezzi-Crivellari and Ritort2016; Vologodskii and Frank-Kamenetskii, Reference Vologodskii and Frank-Kamenetskii2018). Instead, stacking interactions (which are not directly related to the GC-content; Kilchherr et al., Reference Kilchherr, Wachauf, Pelz, Rief, Zacharias and Dietz2016) can play an even more important role in DNA melting than base pairing interactions (Huguet et al., Reference Huguet, Bizarro, Forns, Smith, Bustamante and Ritort2010; Camunas-Soler et al., Reference Camunas-Soler, Ribezzi-Crivellari and Ritort2016; Vologodskii and Frank-Kamenetskii, Reference Vologodskii and Frank-Kamenetskii2018). However, DNA melting under torsion is likely to be even more complex than DNA unzipping, and might depend on additional sequence-dependent molecular features such as DNA flexibility (Vlijm et al., Reference Vlijm, Torre and Dekker2015; Shepherd et al., Reference Shepherd, Greenall, Probert Matt, Noy and Leake Mark2020).
DNA minicircles offer an attractive platform for studying supercoiling-induced DNA denaturation at the molecular level. DNA minicircles consist of small (few hundreds of base pairs) closed DNA molecules that are subjected to high bending and torsional stress. Computer simulations have revealed a rich structural diversity in DNA minicircles, including kinks, base flipping, and denaturation events (Lankaš et al., Reference Lankaš, Lavery and Maddocks2006; Irobalieva et al., Reference Irobalieva, Fogg, Catanese, Sutthibutpong, Chen, Barker, Ludtke, Harris, Schmid, Chiu and Zechiedrich2015; Sutthibutpong et al., Reference Sutthibutpong, Matek, Benham, Slade, Noy, Laughton, doye JP, Louis and Harris2016; Pyne et al., Reference Pyne, Noy, Main, Velasco-Berrelleza, Piperakis, Mitchenall, Cugliandolo, Beton, Stevenson, Hoogenboom, Bates, Maxwell and Harris2020). In a recent study, Sutthibutpong et al. combined atomistic MD, coarse-grained simulations and statistical mechanics calculations to study sequence-dependent melting in DNA minicircles (Sutthibutpong et al., Reference Sutthibutpong, Matek, Benham, Slade, Noy, Laughton, doye JP, Louis and Harris2016). Coarse-grained techniques identified AT-rich regions to be more prone to undergo melting. Nevertheless, detailed atomistic MD simulations indicated that both breathing and melting events were more frequent in flexible pyrimidine-purine dinucleotides such as TpA or CpA (see Section ‘TpA dinucleotides are highly flexible’).
The complex interplay between base pairing, base stacking interactions and DNA flexibility in DNA unwinding remains an open question. We believe that the combination of MD simulations and high-resolution AFM imaging on DNA minicircles can be an extremely useful approach toward gaining such molecular understanding of sequence-dependent DNA melting. Ongoing research in this direction is already yielding very promising results (Pyne et al., Reference Pyne, Noy, Main, Velasco-Berrelleza, Piperakis, Mitchenall, Cugliandolo, Beton, Stevenson, Hoogenboom, Bates, Maxwell and Harris2020).
Chemical modifications and DNA mechanics
In the previous section, we have discussed how the nucleotide sequence impacts the flexibility of canonical Watson–Crick base paired DNA molecules. Inside the cell, however, the DNA often presents changes in its chemical structure, including modified bases, mismatches, or abasic sites. These chemical modifications can occur in the form of epigenetic marks via controlled action of the cellular machinery; or in the form of DNA lesions that jeopardize the normal functioning of the cell.
In this section, we review recent findings on the effects of epigenetics and DNA mismatches on DNA mechanics. When put together, these findings reveal that such chemical modifications have a more complex effect on DNA flexibility than previously thought. Methylation marks play a versatile role in DNA flexibility attending to the sequence context on which they occur and the mechanical deformation considered. On the other hand, DNA mismatches usually enhance DNA flexibility, but confer the DNA with exotic mechanical properties that remain to be deciphered. The emerging picture is that, in both cases, DNA flexibility might act as a potent signal for downstream events. Changes in DNA flexibility resulting from epigenetic marks might affect the compaction state of chromatin; whereas DNA defects might act as flexibility antennas for the recruitment of the repair machinery.
The impact of cytosine methylation on DNA physical properties
Epigenetic DNA marks play a myriad of roles in development (Smith and Meissner, Reference Smith and Meissner2013), aging (Unnikrishnan et al., Reference Unnikrishnan, Freeman, Jackson, Wren, Porter and Richardson2019), and the onset and progression of cancer (Esteller, Reference Esteller2008). The most common of these marks consists of the addition of a methyl group to the C5 carbon of cytosine, which typically occurs at CpG steps. The resulting methylcytosine is usually linked to gene silencing, but the molecular mechanism responsible for this is not completely understood (Cortini et al., Reference Cortini, Barbi, Caré, Lavelle, Lesne, Mozziconacci and Victor2016). Notably, besides affecting the interaction of DNA with several proteins, cytosine methylation is suspected to orchestrate rearrangements of nucleosomes via changes in the mechanical properties of DNA (Dantas Machado, Reference Dantas Machado, Zhou, Rao, Goel, Rastogi, Lazarovici, Bussemaker and Rohs2014; Cortini et al., Reference Cortini, Barbi, Caré, Lavelle, Lesne, Mozziconacci and Victor2016).
Cytosine methylation is generally considered to reduce DNA flexibility Cortini et al. (Reference Cortini, Barbi, Caré, Lavelle, Lesne, Mozziconacci and Victor2016), as supported by recent single-molecule cyclization experiments. Ngo et al. (Reference Ngo, Yoo, Dai, Zhang, He, Aksimentiev and Ha2016) reported that upon methylation of one to eight (four in each strand) cytosines of an arbitrary DNA sequence, the looping time increases, which reflects a decrease in flexibility. This methylation-induced stiffening was well reproduced in supporting MD simulations, showing that methylcytosine dampened local fluctuations in the duplex. In addition, Ngo et al. (Reference Ngo, Yoo, Dai, Zhang, He, Aksimentiev and Ha2016) combined OT and smFRET to show that methylation of the 601 Widom sequence resulted in mechanical destabilization of nucleosomes.
The idea that methylation stiffens the DNA is generally well accepted (Pérez et al., Reference Pérez, Castellazzi, Battistini, Collinet, Flores, Deniz, Ruiz, Torrents, Eritja, Soler-López and Orozco2012; Portella et al., Reference Portella, Battistini and Orozco2013; Cortini et al., Reference Cortini, Barbi, Caré, Lavelle, Lesne, Mozziconacci and Victor2016). Yet, simulation (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a; Liebl and Zacharias, Reference Liebl and Zacharias2018) and experimental studies (Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017; Shon et al., Reference Shon, Rah and Yoon2019) have indicated that, in some cases, methylation can soften the DNA. In particular, methylation could enhance the flexibility of CGIs (discussed in Section ‘CpG islands are rigid DNA regions depleted of nucleosomes’) (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a; Shon et al., Reference Shon, Rah and Yoon2019; Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017). Dense methylation (hypermethylation) of CGIs is a potent gene silencing mechanism, and a hallmark of many cancer types (Esteller, Reference Esteller2008; Deaton and Bird, Reference Deaton and Bird2011). A possible role of DNA flexibility has been suggested in this process (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a; Shon et al., Reference Shon, Rah and Yoon2019; Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017). In fact, CGIs appear rigid, a feature that might explain the nucleosome-binding deficient character of these sequences in vivo (see Section ‘CpG islands are rigid DNA regions depleted of nucleosomes’) (Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017; Shon et al., Reference Shon, Rah and Yoon2019). However, upon hypermethylation, CGIs have been reported to become more flexible, with values of persistence length and stretch modulus closer to those of standard DNA (Pongor et al., Reference Pongor, Bianco, Ferenczy, Kellermayer and Kellermayer2017; Marin-Gonzalez, Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019a; Shon et al., Reference Shon, Rah and Yoon2019). This softening upon hypermethylation could increase the affinity of CGI to form nucleosomes, which may occlude the DNA to the transcription machinery, resulting in gene inactivation. Therefore, despite methylation is usually thought to stiffen the DNA, the full story might be more complex. Given the key biological function of CGIs, it would be of utmost interest to determine whether these sequences constitute an exception to the aforementioned rule.
Moreover, it is worth mentioning that cytosine methylation can have a substantial impact on DNA physical properties other than flexibility (see Table 3). For example, cytosine methylation affects force-induced DNA strand separation, as probed by single-molecule force spectroscopy measurements (Severin et al., Reference Severin, Zou, Gaub and Schulten2011). In this case, cytosine methylation plays a versatile role in DNA mechanics depending on the sequence-context: one methylated cytosine destabilized the duplex but three methylated cytosines resulted in larger mechanical stability. This versatility also appears to be present in sequence-dependent effects of cytosine methylation on DNA structure, as predicted from computer simulations (Pérez et al., Reference Pérez, Castellazzi, Battistini, Collinet, Flores, Deniz, Ruiz, Torrents, Eritja, Soler-López and Orozco2012; Dantas Machado et al., Reference Dantas Machado, Zhou, Rao, Goel, Rastogi, Lazarovici, Bussemaker and Rohs2014; Rao et al., Reference Rao, Chiu, Kribelbauer, Mann, Bussemaker and Rohs2018). For example, in some sequence contexts, cytosine methylation would greatly affect the structure of the DNA, whereas in other sequences the structural effect of this epigenetic mark would be minimal (Rao et al., Reference Rao, Chiu, Kribelbauer, Mann, Bussemaker and Rohs2018). In the extreme case of poly(dG–dC) sequences, single-molecule experiments showed that cytosine methylation can even facilitate the formation of non-canonical Z-DNA structures in the presence of torsional stress (Lee et al., Reference Lee, Kim, Kim and Hong2010a).
Table 3. Effect of cytosine methylation on DNA physical properties

a Single-molecule experiments.
b Computer simulations.
Finally, cytosine methylation has also been studied in the context of DNA condensation by polycations, the phenomenon by which highly positively charged ions mediate DNA compaction by stabilizing inter-helical interactions (Bloomfield, Reference Bloomfield1997). smFRET, MT, and MD simulations coincide that cytosine methylation enhances DNA condensation (Yoo et al., Reference Yoo, Kim, Aksimentiev and Ha2016; Kang et al., Reference Kang, Yoo, Sohn, Lee, Lee, Ma, Kee, Aksimentiev and Kim2018; Yang et al., Reference Yang, Dong, Qiang, Fu, Zhou, Zhang, Yin, Chen, Jia, Dai, Tan and Zhang2020). Importantly, this effect appears consistent when using different condensing polycations (spermidine3+, CoHex3+, spermine4+, and polylysine6+), and DNA sequences.
Altogether, some general conclusions can be derived concerning the effect of cytosine methylation on DNA physical properties. Methylation usually reduces DNA flexibility and facilitates DNA condensation. Nevertheless, in many instances the role of cytosine methylation in DNA mechanics appears strongly dependent on the sequence context on which this epigenetic mark occurs. Therefore, methylation would add an additional layer of complexity to the sequence-dependent biophysical properties of DNA, instead of exerting a systematic, generalizable effect. Understanding cytosine methylation in the context of DNA mechanics would then require investigation on a case-by-case basis.
The effect of DNA mismatches on DNA mechanics
DNA defects, such as mismatches, can induce strongly distorted DNA conformations. Inside the cell, such DNA defects are localized and repaired by the coordinated action of DNA repair proteins with an impressive efficiency. However, the mechanisms by which some of these proteins rapidly identify single DNA imperfections in a huge genome are not completely understood. Remarkably, several DNA repair proteins spanning diverse repair pathways have been reported to interact with a sharply bent DNA (Roberts and Cheng, Reference Roberts and Cheng1998; Natrajan et al., Reference Natrajan, Lamers, Enzlin, Winterwerp, Perrakis and Sixma2003; Qi et al., Reference Qi, Spong, Nam, Banerjee, Jiralerspong, Karplus and Verdine2009; Chakraborty et al., Reference Chakraborty, Steinbach, Paul, Mu, Broyde, Min and Ansari2017; Craggs et al., Reference Craggs, Sustarsic, Plochowietz, Mosayebi, Kaju, Cuthbert, Hohlbein, Domicevica, Biggin, Doye Jonathan and Kapanidis2019; Paul et al., Reference Paul, Mu, Zhao, Ouerfelli, Jeffrey, Broyde and Min2019). A natural question is how (and to which extent) the deformability of damaged DNA impacts these repair processes and, in particular, the recognition of the defect (Krokan and Bjørås, Reference Krokan and Bjørås2013; Marteijn et al., Reference Marteijn, Lans, Vermeulen and Hoeijmakers2014; Kunkel and Erie, Reference Kunkel and Erie2015). For example, do DNA defects spontaneously bend the duplex to recruit repair proteins or do these defects facilitate DNA bending once the protein is attached?
The answer to this question will greatly vary attending to the particular DNA defect and repair proteins involved. Here, we will discuss the effect of mismatches, or ‘pairing’ between non-complementary bases, which have recently received special attention in the context of DNA flexibility (Rossetti et al., Reference Rossetti, Dans, Gomez-Pinto, Ivani, Gonzalez and Orozco2015; Jeong and Kim, Reference Jeong and Kim2019). Mismatches arise during DNA replication or DNA exposure to damaging agents, among other processes. Although these defects are generally associated with increased DNA bendability (see e.g. Vafabakhsh and Ha, Reference Vafabakhsh and Ha2012; Satange et al., Reference Satange, Chang and Hou2018), this might not always be the case, as suggested by recent studies (Rossetti et al., Reference Rossetti, Dans, Gomez-Pinto, Ivani, Gonzalez and Orozco2015; Jeong and Kim, Reference Jeong and Kim2019). Using MD simulations and nuclear magnetic resonance experiments, Orozco, Gonzalez, and coworkers performed a systematic study of the effect of mismatches on DNA flexibility (Rossetti et al., Reference Rossetti, Dans, Gomez-Pinto, Ivani, Gonzalez and Orozco2015). They observed that, in general, the magnitude of bending fluctuations was similar in mismatched duplexes and in control ones with correct base pairing, suggesting that mismatches do not enhance DNA bending flexibility. Nevertheless, DNA molecules containing mismatches were flexible at a local level, with frequent breathing events and distortions of the DNA grooves. The authors warn against simplistic interpretations and indicate that the aforementioned effects are largely dependent on the kind of mismatch considered. Nevertheless, their results suggest that, even though mismatches might not cause spontaneous DNA bending, local DNA distortions due to mismatches may aid proteins at bending the duplex.
Interestingly, Orozco, Gonzalez, and coworkers also observed that DNA distortions induced by defects not only occurred at the position of the mismatch, but also at remote locations of ~4 bp from the mismatch. This finding implies that mismatches might take advantage of a phenomenon known as DNA allostery (Kim et al., Reference Kim, Broströmer, Xing, Jin, Chong, Ge, Wang, Gu, Yang, Gao, Su, Sun and Xie2013) to propagate distortions along the duplex. An independent, experimental observation of such long-range effects of mismatches has been recently reported in the context of single-molecule DNA cyclization (Jeong and Kim, Reference Jeong and Kim2019). Jeong et al. reported that, paradoxically, mismatches destabilize DNA loops, even though these defects favor the sharply bent DNA conformations that are required for loop formation. Such destabilization of loops was attributed to allostery effects, similar to the ones described in Rossetti et al. (Reference Rossetti, Dans, Gomez-Pinto, Ivani, Gonzalez and Orozco2015), but reaching even longer distances of ~50 bp.
Altogether, the appealing hypothesis that mismatches propagate deformations along the DNA and, thus, act as ‘flexibility antennas’ for DNA repair proteins will require further research (see Fig. 3). Alternative mechanisms for the cellular machinery to identify mismatches should also be explored, such as the recently reported finding that mismatches can localize plectoneme tips (Ganji et al., Reference Ganji, Kim, Van Der Torre, Abbondanzieri and Dekker2016; Dittmore et al., Reference Dittmore, Brahmachari, Takagi, Marko and Neuman2017) (see Fig. 3). Note that these two mechanisms, DNA allostery and plectoneme localization, are not necessarily mutually exclusive. Finally, it remains to be explored whether these and other new phenomena arise from other kinds of DNA defects, such as oxo-guanines, photoproducts, or interstrand crosslinks.

Fig. 3. Effects of DNA mismatches on DNA mechanical properties. A single mismatched base pair (center; PDB: 1ONM; Sanchez et al., Reference Sanchez, Volk, Gorenstein and Lloyd2003) can propagate a mechanical signal through the DNA via an allosteric mechanism (left); and can pinpoint the position of a plectoneme tip (right).
Mechanical properties of dsRNA: unexpected differences with dsDNA
Single-molecule studies have also addressed the mechanical properties of dsRNA (Abels et al., Reference Abels, Moreno-Herrero, van der Heijden, Dekker and Dekker2005; Herrero-Galán et al., Reference Herrero-Galán, Fuentes-Perez, Carrasco, Valpuesta, Carrascosa, Moreno-Herrero and Arias-Gonzalez2013; Lipfert et al., Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014; Fu et al., Reference Fu, Zhang, Qiang, Yang, Dai, Tan and Zhang2020). RNA is known to be predominantly single-stranded; however, dsRNA helices are also commonly found inside the cell and exert a myriad of biological functions. For example, dsRNA is the carrier of genetic information in some viruses and dsRNA helices are key elements of tertiary RNA structures, as formed e.g. by ribosomal RNA or t-RNAs (Carter et al., Reference Carter, Clemons, Brodersen, Morgan-Warren, Wimberly and Ramakrishnan2000; Nissen et al., Reference Nissen, Ippolito, Ban, Moore and Steitz2001; Schimmel, Reference Schimmel2018). The mechanical properties of dsRNA might play a role in some of these biological systems. For example, the dsRNA bending stiffness is expected to affect the energetics of genome compaction in dsRNA viruses (Zhang et al., Reference Zhang, Ding, Yu, Chang, Sun and Hong Zhou2015; Buzón et al., Reference Buzón, Maity and Roos2020). Moreover, the sequence-dependent structure and flexibility of dsRNA helices have been proposed to impact dsRNA:protein interactions or the folding of tertiary RNA structures (Perona and Hou, Reference Perona and Hou2007; Yesselman et al., Reference Yesselman, Denny, Bisaria, Herschlag, Greenleaf and Das2019). Finally, a quantitative understanding of dsRNA mechanics could aid the future design of RNA nanostructures (Guo, Reference Guo2010).
Initial studies on the dsRNA persistence length suggested that the mechanical properties of this molecule were qualitatively similar to dsDNA (Hagerman, Reference Hagerman1997; Abels et al., Reference Abels, Moreno-Herrero, van der Heijden, Dekker and Dekker2005). In a seminal study that combined MT and AFM, Abels et al. (Reference Abels, Moreno-Herrero, van der Heijden, Dekker and Dekker2005) obtained a value of P RNA ~62 nm, which is only 20% larger than P DNA. This slightly larger rigidity of dsRNA can be easily rationalized on the basis of the thicker and more compact structure of the A-RNA helix compared to the B-DNA helix. In parallel, MD simulations indicated some differences in the dynamics of dsDNA and dsRNA at the microscopic level (Noy et al., Reference Noy, Pérez, Lankas, Javier Luque and Orozco2004). Namely, dsRNA showed simple deformability patterns that could be well described by few essential motions, whereas dsDNA was able to explore a wider range of conformations. Nonetheless, it was not clear whether these microscopic motions would translate into changes in the global flexibility of the molecules that could be measured in single-molecule experiments.
In the last few years, a number of single-molecule and simulation studies have revealed that the mechanical properties of dsDNA and dsRNA are more different than previously thought (Table 4). The emerging picture is that previous findings on dsDNA flexibility do not necessarily apply to dsRNA. Namely, the latter has its own mechanical identity. In the following, we discuss some of those studies. First, we discuss differences in the flexibility of DNA and RNA duplexes of arbitrary sequence under standard ionic conditions (Sections ‘Stretching flexibility of dsDNA and dsRNA’ and ‘The opposite twist–stretch coupling of dsDNA and dsRNA’). We then comment on the opposite effect of certain multivalent ions on the mechanics of DNA and RNA duplexes (Section ‘The different dynamics of plectoneme formation’). Finally, we discuss sequence effects on the structure (Section ‘Opposite effects of complex ions on the mechanics of dsDNA and dsRNA’) and the flexibility (Section ‘Sequence determinants of intrinsic bending’) of dsDNA and dsRNA.
Table 4. Differences in the mechanical properties of dsDNA and dsRNA

a Single-molecule experiments.
b Computer simulations.
Stretching flexibility of dsDNA and dsRNA
The first single-molecule study that reported qualitative differences in the mechanics of dsDNA and dsRNA was Herrero-Galán et al. (Reference Herrero-Galán, Fuentes-Perez, Carrasco, Valpuesta, Carrascosa, Moreno-Herrero and Arias-Gonzalez2013). The authors first performed AFM and MT experiments on dsDNA and dsRNA and found similar values of the persistence length to the ones previously published in Abels et al. (Reference Abels, Moreno-Herrero, van der Heijden, Dekker and Dekker2005). Nonetheless, OT stretching experiments unveiled an important difference between the two nucleic acids: dsRNA was very soft to stretching deformations, around threefold more flexible compared to dsDNA (see Fig. 4a).
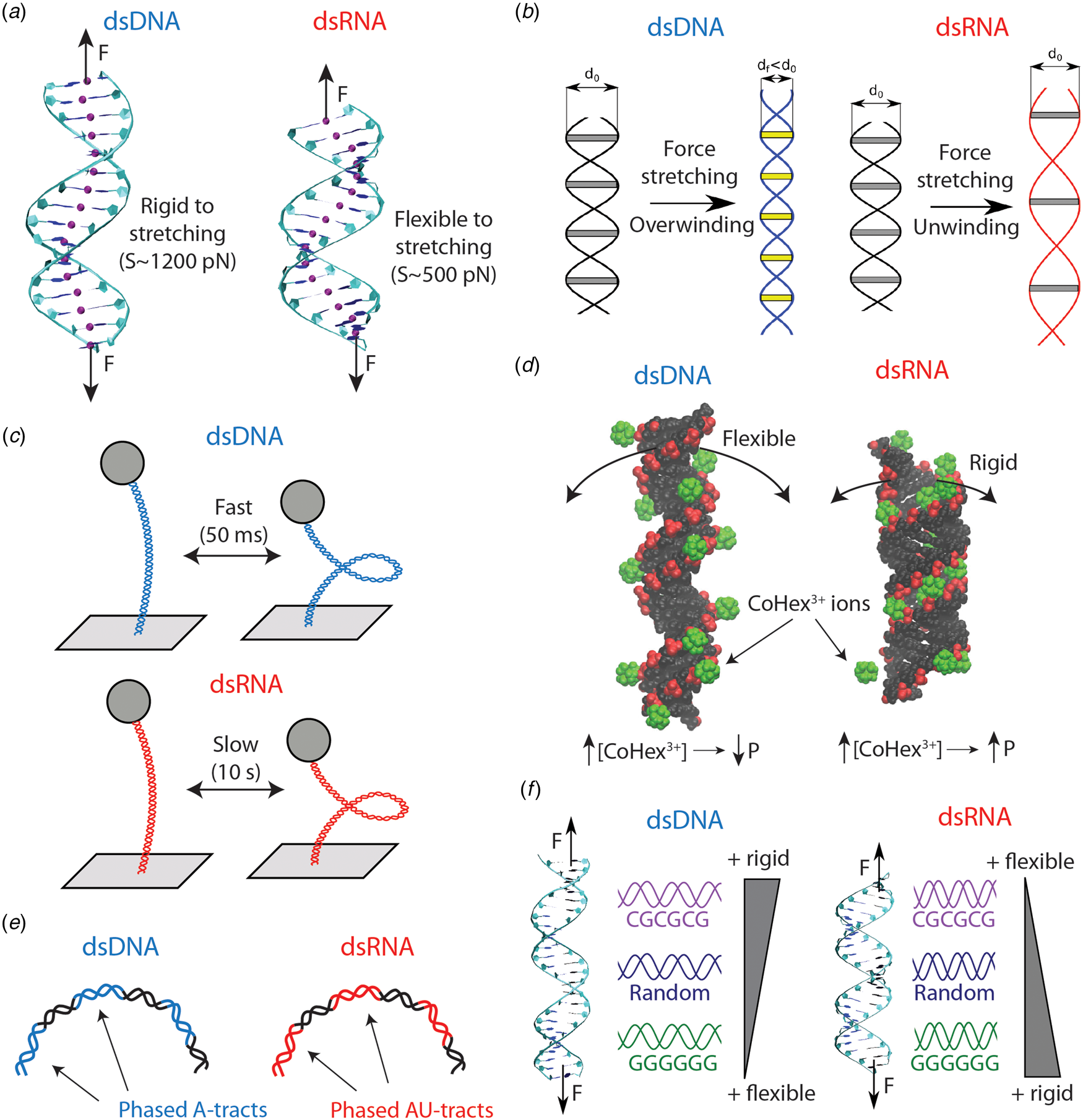
Fig. 4. Differences in the mechanical properties of dsDNA and dsRNA (see also Table 4). (a) dsRNA is around three times softer to stretching deformations than dsDNA. This difference can be explained on the basis of the more open structure of dsRNA, as evidenced from the base pair center chains of the duplexes (purple beads). (b) dsDNA overwinds when stretched, whereas dsRNA unwinds. The peculiar behavior of dsDNA can be rationalized from the shrinking of its radius upon elongation. On the contrary, the dsRNA radius is unchanged when the molecule is stretched. Adapted from Marin-Gonzalez et al. (Reference Marin-Gonzalez, Vilhena, Perez and Moreno-Herrero2017). (c) When the duplexes are supercoiled at the threshold for plectoneme formation, dsDNA displays fast buckling dynamics (ms), whereas dsRNA undergoes slow (s) buckling transitions. This can be partly attributed to the larger persistence length of dsRNA; however, the precise mechanisms for this difference remain incompletely understood. (d) Increasing ionic concentrations result in larger dsDNA bending flexibility and a decrease in persistence length (P). However, some multivalent ions, such as CoHex3+ or spermine4+, can have the opposite effect on dsRNA and stiffen this duplex increasing P. This phenomenon can be understood from the shape of the grooves of the duplexes. For dsRNA, the ions can bind inside the major groove, but in the dsDNA case the ions bind mostly externally. Adapted from Drozdetski et al. (Reference Drozdetski, Tolokh, Pollack, Baker and Onufriev2016). (e) Sequence-induced bending in dsDNA and dsRNA occur, respectively, via A-tract and AU-tract (alternating adenines and uracil) sequences. (f) The nucleotide sequence impacts in different ways the global mechanical properties of dsDNA and dsRNA. Poly(dC–dG) motifs are rigid in dsDNA, but flexible in the dsRNA case; whereas poly(dG:dC) ones are flexible in dsDNA but rigid in dsRNA. Adapted from Marin-Gonzalez et al. (Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019b).
This different stretching flexibility of dsDNA and dsRNA has been reproduced in several experimental and computational studies (Chou et al., Reference Chou, Lipfert and Das2014; Lipfert et al., Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014; Bao et al., Reference Bao, Zhang, Shi, Wu and Tan2017; Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Perez and Moreno-Herrero2017; Fu et al., Reference Fu, Zhang, Qiang, Yang, Dai, Tan and Zhang2020). At the molecular level, the softer stretching response of dsRNA is commonly associated with the more open structure of the A-RNA helix compared to B-DNA (Chou et al., Reference Chou, Lipfert and Das2014; Bao et al., Reference Bao, Zhang, Shi, Wu and Tan2017; Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Perez and Moreno-Herrero2017). Note that this idea is similar to the one presented in Section ‘Poly(dA–dT) and poly(dG:dC) adopt A-like DNA conformations’, according to which DNA sequences with larger A-philicity would possess a softer stretching rigidity.
The opposite twist–stretch coupling of dsDNA and dsRNA
Measurements of the torsional response of dsDNA and dsRNA, as performed by Lipfert et al. revealed more striking differences in the mechanics of these duplexes (Lipfert et al., Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014). Note that when a double-helix is straightened, two parallel strands reach a longer extension than they have when coiled around each other in a helical conformation. According to this picture, stretching a double-helix should lead to unwinding. However, it was known that dsDNA had the surprising ability of overwinding when stretched (Gore et al., Reference Gore, Bryant, Nöllmann, Le, Cozzarelli and Bustamante2006a; Lionnet et al., Reference Lionnet, Joubaud, Lavery, Bensimon and Croquette2006) (see Fig. 4b). Using MT, Lipfert et al. measured changes in the dsRNA extension as a function of twist, and they found the completely opposite behavior: contrary to dsDNA, dsRNA unwinds upon stretching Lipfert et al. (Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014).
The opposite twist–stretch coupling of dsDNA and dsRNA resulted in a great challenge for computational models (see e.g. the review by Kriegel et al., Reference Kriegel, Ermann and Lipfert2017a). Several coarse-grain models, with varying levels of detail, unsuccessfully attempted to reproduce the opposite twist–stretch coupling of dsDNA and dsRNA (Kriegel et al., Reference Kriegel, Ermann and Lipfert2017a). For example, a base pair level model built from crystal structures of dsDNA and dsRNA concluded that both molecules would unwind when stretched (Chou et al., Reference Chou, Lipfert and Das2014). The oxDNA/oxRNA models, which were originally designed to reproduce the thermodynamics of duplex formation (base pairing and stacking interactions), yielded the correct twist–stretch coupling for dsRNA, but failed for dsDNA (Matek et al., Reference Matek, Ouldridge, Doye and Louis2015a, Reference Matek, Šulc, Randisi, Doye and Louis2015b).
The shortfall of coarse-grain simulations suggested that, in order to reproduce the opposite twist–stretch coupling of dsDNA and dsRNA, fine details of the dynamics of the duplexes might need to be considered. Atomistic MD simulations met this demanding condition, as was elegantly demonstrated by Liebl et al. (Reference Liebl, Drsata, Lankas, Lipfert and Zacharias2015). In that study, the authors performed atomistic MD simulations of a dsDNA and a dsRNA molecule of analogous sequence. An analysis of the MD trajectories revealed that the correlation between elongation and twisting motions were of opposite sign for both duplexes, in semi-quantitative agreement with the experiments. The authors further corroborated this finding by exerting a controlled torque on the nucleic acids and measuring the associated change in extension. Consistently, they found that a positive torque (overwinding) caused lengthening of dsDNA, but shortening of dsRNA; and unwinding resulted in the opposite behavior (shortening of dsDNA and lengthening of dsRNA). Liebl et al. elaborated a technical explanation for the opposite twist–stretch coupling of dsDNA and dsRNA based on a wealth of structural parameters of the duplexes. Another study also addressed the twist–stretch coupling of DNA and RNA duplexes and attributed their difference to the slide and inclination motions of the base pairs (Bao et al., Reference Bao, Zhang, Shi, Wu and Tan2017).
Recently, an alternative mechanism for the twist–stretch coupling has been proposed, by directly computing the radii of the duplexes from MD simulations (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Perez and Moreno-Herrero2017) (see Fig. 4b). DNA and RNA duplexes were stretched using a novel constant force protocol for MD simulations Liebl et al. (Reference Liebl, Drsata, Lankas, Lipfert and Zacharias2015). It was found that, as the force increased, dsDNA overwound, while dsRNA unwound, in agreement with the aforementioned simulations and experiments. Importantly, because the molecules were left to equilibrate under stress, it was possible to measure how their average structure changed at each value of the external force. It was thus observed that the dsDNA radius decreases with force, whereas the dsRNA radius remains approximately constant. The force-induced shrinking of the DNA radius brings together the strands of the duplex, allowing it to overwind upon stretching (see Fig. 4b). Since this capability of reducing the radius was absent in dsRNA, this molecule could only unwind when stretched. The larger flexibility of the DNA sugar was suggested to be responsible for promoting the reduction of the dsDNA radius.
The different dynamics of plectoneme formation
The extensive investigation of Lipfert et al. on the twisting dynamics of dsDNA and dsRNA yielded yet another surprising result (Lipfert et al., (Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014). When these double-stranded nucleic acids are twisted above a certain threshold, they undergo a buckling transition and start forming plectonemes (see Fig. 4c). Lipfert et al. (Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014) studied the dynamics of dsDNA and dsRNA when the twist of duplexes is constrained at the threshold value for the buckling transition. At that point, pre-buckling and post-buckling states are equally populated and the nucleic acids continuously transition between states. The authors found that such buckling dynamics were much slower in dsRNA compared to dsDNA, with a difference in dwell times of at least two orders of magnitude.
This issue has been recently addressed in an insightful simulation study. Ott et al. (Reference Ott, Martini, Lipfert and Gerland2020) modeled a dsDNA and a dsRNA molecule using Brownian dynamics simulations and a simple WLC model that contemplates bending and twisting deformations. They found that the difference in persistence length of dsDNA and dsRNA, although small, can result in an order of magnitude change in the buckling dynamics. This approach thus partially explains the experimental findings. However, further theoretical efforts are needed to arrive at a quantitative description of the buckling dynamics of these nucleic acids (see the recent perspective by Lankaš, Reference Lankaš2020). We believe that such description should take into account the cross-talk between twisting and bending deformations, that is, the fact that twisting can facilitate bending. This so-called twist–stretch coupling parameter has recently been estimated for dsDNA (Nomidis et al., Reference Nomidis, Kriegel, Vanderlinden, Lipfert and Carlon2017), but a solid measurement is still lacking in the dsRNA case.
Opposite effects of complex ions on the mechanics of dsDNA and dsRNA
The effect of salt conditions on dsDNA mechanics has been extensively studied for several decades (Harrington, Reference Harrington1978; Ha and Thirumalai, Reference Ha and Thirumalai2003). However, recent studies are providing very interesting insights into this topic (Brunet et al., Reference Brunet, Tardin, Salome, Rousseau, Destainville and Manghi2015b; Kriegel et al., Reference Kriegel, Ermann, Forbes, Dulin, Dekker and Lipfert2017b; Guilbaud et al., Reference Guilbaud, Salomé, Destainville, Manghi and Tardin2019). Nonetheless, in contrast to DNA, much less is known about the effect of salt on dsRNA flexibility. Initial single-molecule experiments reported a qualitatively similar decrease in P RNA and P DNA with increasing monovalent salt (Herrero-Galán et al., Reference Herrero-Galán, Fuentes-Perez, Carrasco, Valpuesta, Carrascosa, Moreno-Herrero and Arias-Gonzalez2013; Lipfert et al., Reference Lipfert, Skinner, Keegstra, Hensgens, Jager, Dulin, Köber, Yu, Donkers, Chou, Das and Dekker2014). A priori, these findings could suggest a similar dependence of DNA and dsRNA flexibility on salt conditions. However, further single-molecule experiments and MD simulations have reported unexpected results when multivalent ions are present (Drozdetski et al., Reference Drozdetski, Tolokh, Pollack, Baker and Onufriev2016; Fu et al., Reference Fu, Zhang, Qiang, Yang, Dai, Tan and Zhang2020) (see Fig. 4d). Motivated by the different condensation properties of dsDNA and dsRNA (Li et al., Reference Li, Pabit, Meisburger and Pollack2011; Tolokh et al., Reference Tolokh, Pabit, Katz, Chen, Drozdetski, Baker, Pollack and Onufriev2014), Onufriev and coworkers performed atomistic MD simulations to explore the role of the ion CoHex3+ in P RNA and P DNA (Drozdetski et al., Reference Drozdetski, Tolokh, Pollack, Baker and Onufriev2016). The simulations showed that P DNA decreases with increasing CoHex3+ concentration, in accordance with naïve intuition and with previous experiments (Baumann et al., Reference Baumann, Smith, Bloomfield and Bustamante1997). However, unexpectedly, increasing amounts of CoHex3+ resulted in an increase of P RNA. This stiffening of dsRNA was consistent when alternative multivalent ions were used, such as spermine4+ or a hypothetical Na3+. Recently, the opposite effect of CoHex3+ on the flexibility of dsDNA and dsRNA has been experimentally demonstrated in a comprehensive study that combines both MT experiments and MD simulations (Fu et al., Reference Fu, Zhang, Qiang, Yang, Dai, Tan and Zhang2020). Altogether, these studies illustrate the limitations of polyelectrolyte models to describe the interaction of dsDNA and dsRNA with complex, multivalent ions such as CoHex3+. In these cases, molecular details of the double helices, such as shape of their grooves, must be taken into account in order to explain the effect of these ions on the elasticity of the duplexes.
Sequence determinants of intrinsic bending
The phenomenon of sequence-induced DNA bending by A-tracts raises the question of whether a similar effect can occur in dsRNA. This question is more complicated than one would expect. For example, the few structures of naked RNA duplexes available from X-ray crystallography experiments often present artifacts (Šponer et al., Reference Šponer, Bussi, Krepl, Banáš, Bottaro, Cunha, Gil-Ley, Pinamonti, Poblete, Jurečka, Walter and Otyepka2018). This issue constitutes an important drawback for experimental validation of dsRNA MD simulations via X-ray crystallography data (Šponer et al., Reference Šponer, Bussi, Krepl, Banáš, Bottaro, Cunha, Gil-Ley, Pinamonti, Poblete, Jurečka, Walter and Otyepka2018). Testing MD against single-molecule experiments can therefore be a promising route for exploring sequence-dependent dsRNA features. By combining MD simulations and AFM experiments, it has been recently shown that alternating adenines and uracils – or AU-tracts – bend the RNA duplex (Marin-Gonzalez et al., Reference Marin-Gonzalez, Aicart-Ramos, Marin-Baquero, Martín-González, Suomalainen, Kannan, Vilhena, Greber, Moreno-Herrero and Pérez2020b) (see Fig. 4e). AU-tracts were long known to possess a peculiar structure at a local base pair level (Dock-Bregeon et al., Reference Dock-Bregeon, Chevrier, Podjarny, Johnson, De Bear, Gough, Gilham and Moras1989), but their effect on global dsRNA features had remained largely unexplored. The MD simulations from Marin-Gonzalez et al. (Reference Marin-Gonzalez, Aicart-Ramos, Marin-Baquero, Martín-González, Suomalainen, Kannan, Vilhena, Greber, Moreno-Herrero and Pérez2020b) revealed that dsRNA molecules with AU-tracts were systematically more bent than sequences lacking this motif. Motivated by this finding, dsRNA molecules were fabricated with repetitions of AU-tracts spaced by 11 bp and these molecules were imaged using an AFM. The images revealed a bent character in these AU-tract molecules, which was quantified by a value of the persistence length as low as ~30 nm, about half the standard value of dsRNA (~60 nm). These findings argue against simplistic conceptions of dsRNA as a regular helix. Further investigation will be required to elucidate the molecular mechanisms and the possible biological consequences of this novel phenomenon.
Sequence-dependent mechanical properties
The aforementioned sequence-dependent dsRNA bending demonstrates that the nucleotide sequence can affect in a very distinct manner the structure of dsDNA and dsRNA. Namely, A-tracts induce an intrinsic bend in dsDNA, but not in dsRNA; and AU-tracts have the opposite effect: they bend dsRNA, but not dsDNA (see Fig. 4e). Therefore, it would be natural to expect that the nucleotide sequence would also impact the relative flexibility of dsDNA and dsRNA in different ways. A recent simulation study suggests that this might be the case (Marin-Gonzalez et al., Reference Marin-Gonzalez, Vilhena, Moreno-Herrero and Perez2019b). Using MD simulations, Marin-Gonzalez et al. measured the mechanical response of several dsRNA sequences and compared the results with dsDNA analogs of the same sequence. dsRNA molecules were always more flexible to stretching deformations than dsDNA ones, regardless of the sequence. However, the effect of the nucleotide sequence on the stretching flexibility was completely different in the two nucleic acids. For example, the poly(rG:rC) RNA duplex is relatively rigid to stretching, but a poly(dG:dC) DNA duplex is highly flexible when compared with an arbitrary sequence (see Fig. 4f). On the contrary, the poly(rG–rC) dsRNA molecule is flexible to stretching, but the poly(dG–dC) dsDNA is rigid. The molecular mechanisms behind this difference are to be examined. Furthermore, the idea that sequence effects on dsDNA and dsRNA flexibility can be substantially different still awaits experimental validation.
Conclusions and future perspectives
In this review, we have revisited recent single-molecule experiments and MD simulations studies on DNA mechanical properties. These studies are collectively providing a comprehensive, molecular description of DNA mechanics by assessing how microscopic chemical features of the double-helix impact its physical properties. Importantly, as we deepen into such molecular characterization, a rich sequence-dependent conformational variability of the double-helix emerges, which is often overlooked by classical polymer approaches. For example, both experiments and simulations suggest that, besides few exceptions, the DNA persistence length is relatively insensitive to the nucleotide sequence (Geggier and Vologodskii, Reference Geggier and Vologodskii2010; Mitchell et al., Reference Mitchell, Glowacki, Grandchamp, Manning and Maddocks2017). Even distorted DNA duplexes containing a mismatch appear to have a standard bending flexibility (Rossetti et al., Reference Rossetti, Dans, Gomez-Pinto, Ivani, Gonzalez and Orozco2015). On the contrary, local, strong deformations such as kinks, bubbles or Z-DNA structures are strongly sequence-dependent (Rich and Zhang, Reference Rich and Zhang2003; Olson and Zhurkin, Reference Olson and Zhurkin2011; Fogg et al., Reference Fogg, Randall, Pettitt, Sumners, Harris and Zechiedrich2012). It is thus tempting to conclude that sequence effects on DNA mechanics are amplified under large mechanical stress, that is, when the duplex is forced to adopt a structure that substantially differs from the canonical B-DNA helix.
Furthermore, the studies here reviewed are enabling a better understanding of the mechanical impact of cytosine methylation. Several lines of evidence indicate that cytosine methylation affects many DNA physical properties in a highly complex manner that often depends on the particular sequence context (see Table 3 and reference Cortini et al., Reference Cortini, Barbi, Caré, Lavelle, Lesne, Mozziconacci and Victor2016). The emerging view is that, rather than exerting a systematic effect on DNA mechanics, methylated cytosine acts as a ‘fifth nucleotide’ that expands the ‘physical code’ imprinted in a given DNA sequence. Similar considerations might apply to DNA mismatches, whose effects on DNA mechanics appear to be strongly dependent on the specific kind of mismatch considered.
Finally, we have revisited recent findings on dsRNA mechanics, focusing on those studies that reveal unexpected observations in the mechanical properties of dsRNA when compared to its DNA counterpart. From a molecular perspective, these studies raise an interesting consideration. Namely, that the presence of an extra –OH group in the sugar and the substitution of thymine by uracil have enormous implications in the physical properties of nucleic acids.
Despite the substantial progress made in the last few years, a number of important aspects of DNA mechanics will require further study. In the following, we comment on some of those aspects and we briefly discuss how an improved characterization of DNA physical properties can potentially impact other areas of biology, biophysics, and nanotechnology.
Sequence-dependent DNA mechanics beyond the elastic regime
In the elastic regime, the sequence-dependent DNA deformability can be accurately described from the analysis of structural databases (Olson et al., Reference Olson, Gorin, Lu, Hock and Zhurkin1998), or from extensive atomistic MD simulations (Pasi et al., Reference Pasi, Maddocks, Beveridge, Bishop, Case, Cheatham, Iii Dans, Jayaram, Lankas, Laughton, Mitchell, Osman, Orozco, Pérez, Petkevičiūtė, Spackova, Sponer, Zakrzewska and Lavery2014; Walther et al., Reference Walther, Dans, Balaceanu, Hospital, Bayarri and Orozco2020). However, as mentioned above, the nucleotide sequence largely affects the energetics of highly distorted DNA conformations beyond the elastic regime, such as kinks or bubbles. We have outlined in a qualitative manner the main sequence determinants of such sharp DNA deformations (see Table 2). However, a quantitative characterization of DNA dynamics beyond the elastic regime is a challenge that will need to be addressed in future studies. In this respect, novel high-throughput assays based on next-generation sequencing, such as the recently developed loop-seq assay (Basu et al., Reference Basu, Bobrovnikov, Cieza, Qureshi and Ha2020, Reference Basu, Bobrovnikov, Qureshi, Kayikcioglu, Ngo, Anand, Eustermann, Cieza, Morgan, Hejna, Rube, Hopfner, Wolberger, Song and Ha2021), offer an attractive platform to interrogate such sequence-dependent DNA mechanical properties.
It is important to note that DNA kinks, bubbles, and other distorted conformations appear in a number of structures of DNA:protein complexes, including the nucleosome core (Dickerson et al., Reference Dickerson1998; Olson and Zhurkin, Reference Olson and Zhurkin2011). Therefore, a quantitative understanding of the energetics of these conformations might shed light on several biological questions. In the paradigmatic case of nucleosome stability, DNA kinks might be more determinant than the smooth bending flexibility (Zhurkin and Olson, Reference Zhurkin and Olson2013). Together with other sequence-dependent features, such as DNA shape (Rohs et al., Reference Rohs, West, Sosinsky, Liu, Mann and Honig2009), DNA kinks might greatly contribute to the wide sequence-dependent variability of nucleosome affinity reported in in vitro experiments (Onufriev and Schiessel, Reference Onufriev and Schiessel2019).
The future challenges regarding the sequence-dependent DNA mechanics beyond the elastic regime can be summarized as:
• To quantitatively characterize the energetics of formation of highly distorted DNA conformations, most notably kinks and local denaturation events.
• To devise novel assays for systematically evaluating the sequence-dependent energetics of formation of highly distorted DNA conformations.
• To evaluate under which circumstances the in vivo mechanical stress, as found e.g. in the nucleosome core, is sufficient for stabilizing the formation of those highly distorted DNA structures.
• To incorporate non-elastic effects into current coarse-grain models of DNA to improve theoretical descriptions of DNA mechanics.
Toward a better characterization of cytosine methylation and DNA mismatches in the context of DNA mechanics
As discussed in the text, cytosine methylation is usually correlated with an increased rigidity of the DNA. However, there is evidence pointing toward possible exceptions to this rule, most notably, certain sequences with high density of CpG steps (or CGIs). These (and potentially other kinds of) DNA sequences where methylation results in DNA softening clearly deserve more attention, both for experiments and simulations. The latter should aim to elucidate the molecular mechanisms behind methylation-induced changes in DNA flexibility, which still remain obscure.
In addition, future efforts are needed to better characterize the effect of cytosine methylation on DNA condensation by polycations. This question is particularly timely, in light of recent experiments and simulations that have suggested a liquid–liquid phase separation mechanism for DNA condensation (Kang et al., Reference Kang, Yoo, Sohn, Lee, Lee, Ma, Kee, Aksimentiev and Kim2018; Shakya and King, Reference Shakya and King2018). It is thus conceivable that cytosine methylation might enhance the phase separation behavior of DNA.
Understanding how DNA mismatches alter the DNA physical properties is another exciting task for the near future. For example, the hypothesis that mismatches promote allosteric effects in the DNA would benefit from more extensive experimental support at the single-molecule level. Another interesting idea to test experimentally would be whether different kinds of mismatches possess different mechanical footprints. Testing transductions (non-complementary pyrimidine:purine base pairing) against transversions (pyrimidine:pyrimidine or purine:purine base pairing) would be a promising starting point. Because the geometries of these kinds of mismatches are very different, it is expected they will possess different mechanical properties amenable to experimental observation.
Altogether, we devise the following challenges for future studies on cytosine methylation and DNA mismatches in the context of DNA mechanics:
• To further test experimentally the effect of cytosine methylation on the dynamics of CGIs and explore other potential exceptions to the rule that methylation reduces DNA flexibility.
• To further test the effect of cytosine methylation on the condensation and, potentially, liquid–liquid phase separation behavior of DNA.
• To provide additional experimental evidence on allosteric DNA effects induced by DNA mismatches.
• To explore the variability of mechanical effects among different kinds of mismatches.
DNA and RNA mechanics at the service of nanotechnology
Quoting Richard Feynman, ‘what I cannot create, I do not understand’. True comprehension of natural processes is achieved when those processes can be customized to fulfill our necessities. In the last few decades, the field of DNA nanotechnology has provided an outstanding example of this philosophy. DNA nanostructures can be designed from molecular models that incorporate well-characterized biophysical properties of the DNA, including mechanical parameters such as the persistence length, the twist stiffness, or the twist–stretch coupling (Dietz et al., Reference Dietz, Douglas and Shih2009; Ouldridge et al., Reference Ouldridge, Louis and Doye2010; Castro et al., Reference Castro, Kilchherr, Kim, Shiao, Wauer, Wortmann, Bathe and Dietz2011; Maffeo and Aksimentiev, Reference Maffeo and Aksimentiev2020). Nevertheless, when conceiving DNA as nanomaterial, considerations of sequence effects are often limited to base pairing and stacking interactions (Doye et al., Reference Doye, Ouldridge, Louis, Romano, Šulc, Matek, Snodin, Rovigatti, Schreck, Harrison and Smith2013). Only in few cases, additional sequence-dependent features of dsDNA, such as the B–Z transition or A-tract curvature, have been exploited to achieve novel functionalities of DNA nanodevices, such as molecular switches or curved DNA trajectories (Mao et al., Reference Mao, Sun, Shen and Seeman1999; Iric et al., Reference Iric, Subramanian, Oertel, Agarwal, Matthies, Periole, Sakmar, Huber, Fahmy and Schmidt2018). Incorporating sequence-dependent biophysical properties, such as the ones delineated in Table 2, in the design of future DNA nanodevices could therefore expand the potential of DNA as nanotechnological material. Recent exciting developments in coarse-grain models of DNA mechanics hold great promise for this ambitious goal (Ouldridge et al., Reference Ouldridge, Louis and Doye2010; Edens et al., Reference Edens, Brozik and Keller2012; Freeman et al., Reference Freeman, Hinckley, Lequieu, Whitmer and De Pablo2014; Chakraborty et al., Reference Chakraborty, Hori and Thirumalai2018; Maffeo and Aksimentiev, Reference Maffeo and Aksimentiev2020).
In addition to DNA, the field of RNA nanotechnology is becoming increasingly popular. An improved quantitative comprehension of sequence-dependent dsRNA structure and flexibility will surely accompany a sustained and solid development of the RNA nanotechnology field (Guo, Reference Guo2010; Jasinski et al., Reference Jasinski, Haque, Binzel and Guo2017). Note that the rules that govern dsRNA mechanics and structure are different from those of dsDNA (see Table 4 and references therein). Namely, RNA possesses its own material properties, different from those of DNA, and might therefore offer new, unforeseen possibilities in terms of molecular design. Exploiting the unique biophysical properties of RNA for nanotechnological applications will be an exciting challenge for the years to come.
In conclusion, we foresee the following challenges for expanding the application of nucleic acids in nanotechnology:
• To exploit sequence-dependent structural and mechanical features in the design of DNA-based nanostructures.
• To advance our current knowledge on dsRNA mechanics and exploit these and other biophysical properties of RNA for improved design of RNA-based nanostructures.
Acknowledgements
We thank financial support from the Spanish MINECO (projects MAT2017-83273-R (AEI/FEDER, UE) to R.P., BFU2017-83794-P (AEI/FEDER, UE to F.M.-H.), and Comunidad de Madrid (Tec4Bio – S2018/NMT-4443 and NanoBioCancer – Y2018/BIO-4747 to F.M.-H.). R.P. acknowledges support from the Spanish Ministry of Science and Innovation, through the ‘María de Maeztu’ Programme for Units of Excellence in R&D (CEX2018-000805-M). F.M.-H. acknowledges support from European Research Council (ERC) under the European Union Horizon 2020 research and innovation (grant agreement No 681299). J.G.V. acknowledges funding from a Marie Sklodowska Curie Fellowship within the Horizons 2020 framework (DLV-795286). A.M.-G. acknowledges support from the International PhD Program of ‘La Caixa-Severo Ochoa’ as a recipient of a PhD fellowship.