Introduction
Neurons communicate by releasing neurotransmitters at the synapse, a process driven by the fusion of synaptic vesicles with the plasma membrane (Südhof, Reference Südhof, Südhof and Starke2008). This fusion event, mediated by the fusion machinery, occurs within a submillisecond timescale following an action potential, triggering presynaptic Ca currents which activate fusion (Borst and Sakmann, Reference Borst and Sakmann1998; Südhof, Reference Südhof2013). The transmitter that is released binds to postsynaptic receptors, triggering postsynaptic currents (Südhof, Reference Südhof2013). But what are the components of this machinery, and how do they work together to achieve such rapid fusion?
At the end of the 20th century, experiments identified the core of the fusion machinery as the SNARE complex, composed of Synaptobrevin 2 (Syb2), Syntaxin-1 (Stx1), and SNAP-25 (Baumert et al., Reference Baumert1989; Oyler et al., Reference Oyler1989; Bennett et al., Reference Bennett1992; Sollner et al., Reference Sollner1993), along with several accessory proteins, including Synaptotagmin (Syt), Complexin (Cpx), Munc13, and Munc18 (Perin et al., Reference Perin1990; Hata et al., Reference Hata1993; Brose et al., Reference Brose1995; McMahon et al., Reference McMahon1995) (Figure 1a). in vitro reconstitution experiments demonstrated that Syb2, located on synaptic vesicles (Baumert et al., Reference Baumert1989), and Stx1 and SNAP-25, on the plasma membrane (Oyler et al., Reference Oyler1989; Bennett et al., Reference Bennett1992), self-assemble into a SNARE complex (Fasshauer and Margittai, Reference Fasshauer and Margittai2004). This complex forms the minimal machinery necessary for vesicle fusion (Weber et al., Reference Weber1998). Subsequent studies identified Syt as the calcium sensor that detects Ca2+ influx through voltage-gated calcium channels via its C2 domains, triggering Ca2+-evoked release (Li et al., Reference Li1995; Chapman, Reference Chapman2002) while clamping spontaneous fusion by maintaining vesicle priming (Voleti et al., Reference Voleti2020; Rizo, Reference Rizo2022). Different Syt isoforms serve at different speeds of release upon Ca signals. The fast Ca sensor Syt1 mediates synchronous release, whereas the slow sensor Syt7 was reported to trigger asynchronous release (Bacaj et al., Reference Bacaj2013). Subsequently, Syt7 was found to function in Ca2+-dependent synaptic vesicle replenishment in interaction with calmodulin (Liu et al., Reference Liu2014). Cpx was later recognized as a modulator of neurotransmitter release, both inhibiting spontaneous fusion and enhancing Ca2+-evoked release (Jorquera et al., Reference Jorquera2012; Li et al., Reference Li2024). Additionally, Munc13 and Munc18 facilitate SNARE complex assembly and vesicle docking and priming while competing with their disassembly by N-ethylmaleimide–sensitive factor (NSF) and one of the soluble NSF attachment proteins (α-SNAP) (Ma et al., Reference Ma2013; Liu et al., Reference Liu2016; White et al., Reference White2018; Rizo, Reference Rizo2022).
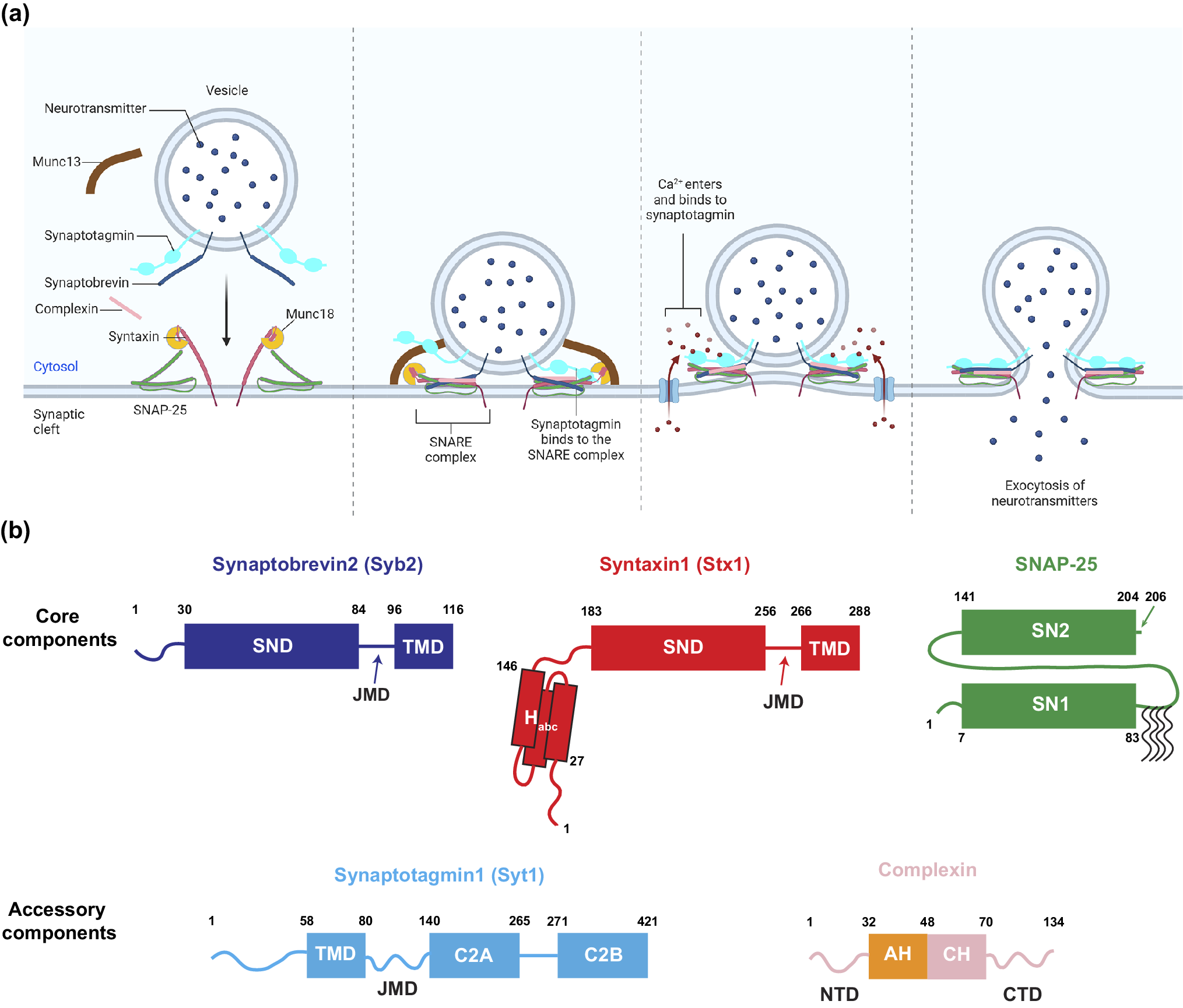
Figure 1. Components of the SNARE-mediated membrane fusion machinery.
(a) The fusion machinery is composed of core SNARE proteins – Syb2, Stx1, and SNAP-25 – along with accessory regulatory proteins such as Syt1, Cpx, Munc13, and Munc18. These components cooperate to rapidly drive neurotransmitter release. (b) Protein Domain Strucures. Syb2 and Stx1 each contain one SND and one TMD, connected by a JMD (Stein et al., Reference Stein2009). Unlike Syb2, which has only an unstructured N-terminal domain (NTD), Stx1 contains an N-terminal Habc domain composed of three helices, connected to the SND via a 37-residue linker. SNAP-25 has two SNARE domains, SN1 and SN2, connected by a 58-residue linker, with several N-terminal residues (positions 85, 88, 90, and 92) that are commonly palmitoylated (Veit et al., Reference Veit1996). The Ca2+ sensor Syt1 is located on synaptic vesicles, with its TMD at the N-terminus. A 60-residue JMD connects the TMD to the C-terminal tandem C2 domains (C2A and C2B), which mediate Ca2+ binding (Fernandez et al., Reference Fernandez2001). Complexin contains several helical domains including the accessory helix (AH) and the central helix (CH), flanked by an NTD and a C-terminal domain (CTD), respectively (Rizo, Reference Rizo2022). All residues at the edge of domains are marked with residue numbers.
Despite extensive experimental studies elucidating the roles of SNARE proteins and their regulators, directly visualizing membrane fusion – how the fusion machinery assembles, reshapes membranes, and drives neurotransmitter release – remains a challenge due to the nanometer-scale dimensions (Jung, Reference Jung2019) and transient kinetics of this process (Sabatini and Regehr, Reference Sabatini and Regehr1996; Acuna et al., Reference Acuna2014). Molecular dynamics (MD) simulations have emerged as a powerful complementary approach, providing details of protein–protein and protein–lipid interactions that are otherwise inaccessible. These simulations not only generate dynamic ‘movies’ of membrane fusion but also offer mechanistic insights that can guide future experiments.
MD simulations offer a choice between different levels of resolution: all-atom (AA) simulations, which model every atom for high accuracy in molecular interactions and motions, and coarse-grained (CG) simulations, which simplify molecular representations to capture large-scale conformational changes over extended timescales (Marrink et al., Reference Marrink2007; Deserno, Reference Deserno2009). These simulations provide a diversity of insight to uncover the mechanism of the fusion machinery to cooperatively and rapidly fuse membranes.
For CG simulations, we mainly discuss two kinds of force fields: The MARTINI force field (Marrink et al., Reference Marrink2004, Reference Marrink2007; Monticelli et al., Reference Monticelli2008) and an ultra-coarse-grain (UCG) force field applied in the O’Shaughnessy group (Mostafavi et al., Reference Mostafavi2017; McDargh et al., Reference McDargh2018; Butu et al., Reference Butu, An and O’Shaughnessy2025). The MARTINI (https://cgmartini.nl/) force field represents a molecule based on a four-to-one mapping scheme (Marrink et al., Reference Marrink2004, Reference Marrink2007; Monticelli et al., Reference Monticelli2008) (About four non-hydrogen atoms and associated hydrogens are represented by a single bead). Some chemical groups, like ring-like compounds, are represented with higher resolution with a bead representing two non-hydrogen atoms and associated hydrogens. Only five main types of interactions, namely polar, non-polar, apolar, charged, and halogen, are defined. The bonded interactions are typically derived from AA simulations, and the non-bonded interactions are tuned based on the reproduction of experimental free energies between polar and apolar phases of chemical compounds. Using the CG MARTINI force field accelerates molecular dynamics like diffusion by a factor of 4 (Marrink et al., Reference Marrink2004, Reference Marrink2007). The UCG representations are much more coarse grained than the MARTINI force field (Mostafavi et al., Reference Mostafavi2017; McDargh et al., Reference McDargh2018; Butu et al., Reference Butu, An and O’Shaughnessy2025). For the SNARE proteins, one bead represents a group of four residues and representations of lipids, adopted from (Illya and Deserno (Reference Illya and Deserno2008), use only four beads to represent a lipid molecule. The solvent molecules are represented implicitly. This UCG method significantly increases the dynamics such that the simulation can easily reach milliseconds.
In this review, we explore how multiscale MD simulations have advanced our understanding of SNARE-mediated membrane fusion. We begin by examining current perspectives on how SNARE complexes generate forces to remodel membranes – whether through SNARE domain (SND) zippering, entropic forces, or a combination of both – by analyzing the roles of SNARE juxtamembrane domains (JMDs) and transmembrane domains (TMDs) (Figure 1b). We then discuss possible pathways leading to fusion pore (FP) formation and the kinetics governing this process. Next, we highlight how MD simulations have provided insights into how Syt1 C2 domains regulate SNARE dynamics to achieve Ca2+ sensing, as well as the potential roles of Cpx in modulating fusion by either clamping or facilitating it under different conditions. We also examine the complementary use of AA and CG simulations, balancing accuracy and computational efficiency to capture both molecular interactions and long-timescale dynamics. Finally, we discuss how AI tools like AlphaFold and Robetta have expanded the scope of MD simulations by improving protein structure predictions, enabling the study of isoform-specific differences, and facilitating the integration of additional fusion machinery components into simulations.
SNARE complex: The core engine of membrane fusion
SNARE zippering and membrane contact formation
Since Weber et al. discovered that SNARE proteins form the minimal membrane fusion machinery (Weber et al., Reference Weber1998), a key question has been how SNARE proteins drive vesicle–plasma membrane fusion. SNARE complexes are formed by 1 R-SND and 3 Q-SNDs whose layer 0 residue at the center of their SNDs are Arginine (R) and Glutamine (Q), respectively (Yadav et al., Reference Yadav2024). Experiments have shown that in the neuronal SNARE complex the R-SNARE Syb2 zippers into the Stx-SNAP25 t-SNARE (Q-SNARE) complex, forming a coiled coil structure (Chen et al., Reference Chen2001). The zippering is exergonic and for the amount of energy released during zippering estimates in the range of 30–85 kT have been reported (Wiederhold and Fasshauer, Reference Wiederhold and Fasshauer2009; Gao et al., Reference Gao2012; Ma et al., Reference Ma2015; Zhang and Hughson, Reference Zhang and Hughson2021; Jahn et al., Reference Jahn2024). However, the roles of the SNDs, JMDs and TMDs in this process remain controversial. Two competing hypotheses for the origins of the SNARE complex driving forces to achieve fusion have emerged: (1) the released energy during SND zippering is transduced to the TMDs via rigid JMDs, bringing the two membranes into close proximity and enabling neurotransmission (Figure 2a) (Südhof and Rothman, Reference Südhof and Rothman2009; Jackson, Reference Jackson2010; Zhang and Hughson, Reference Zhang and Hughson2021), or (2) SNARE complexes bring the membranes together via entropic forces generated by thermodynamic fluctuations of the SNARE domains to enable a larger available space for SND occupancy, whereas the SND zippering energy had been dissipated, transduced by flexible JMDs (Figure 2b) (Mostafavi et al., Reference Mostafavi2017; McDargh et al., Reference McDargh2018; Butu et al., Reference Butu, An and O’Shaughnessy2025). For more detailed discussions of the pathways of SNARE assembly, fusion triggering, and disassembly see (Jahn et al., Reference Jahn2024). In this section, we review and focus primarily on the evidence from MD simulations supporting these hypotheses.
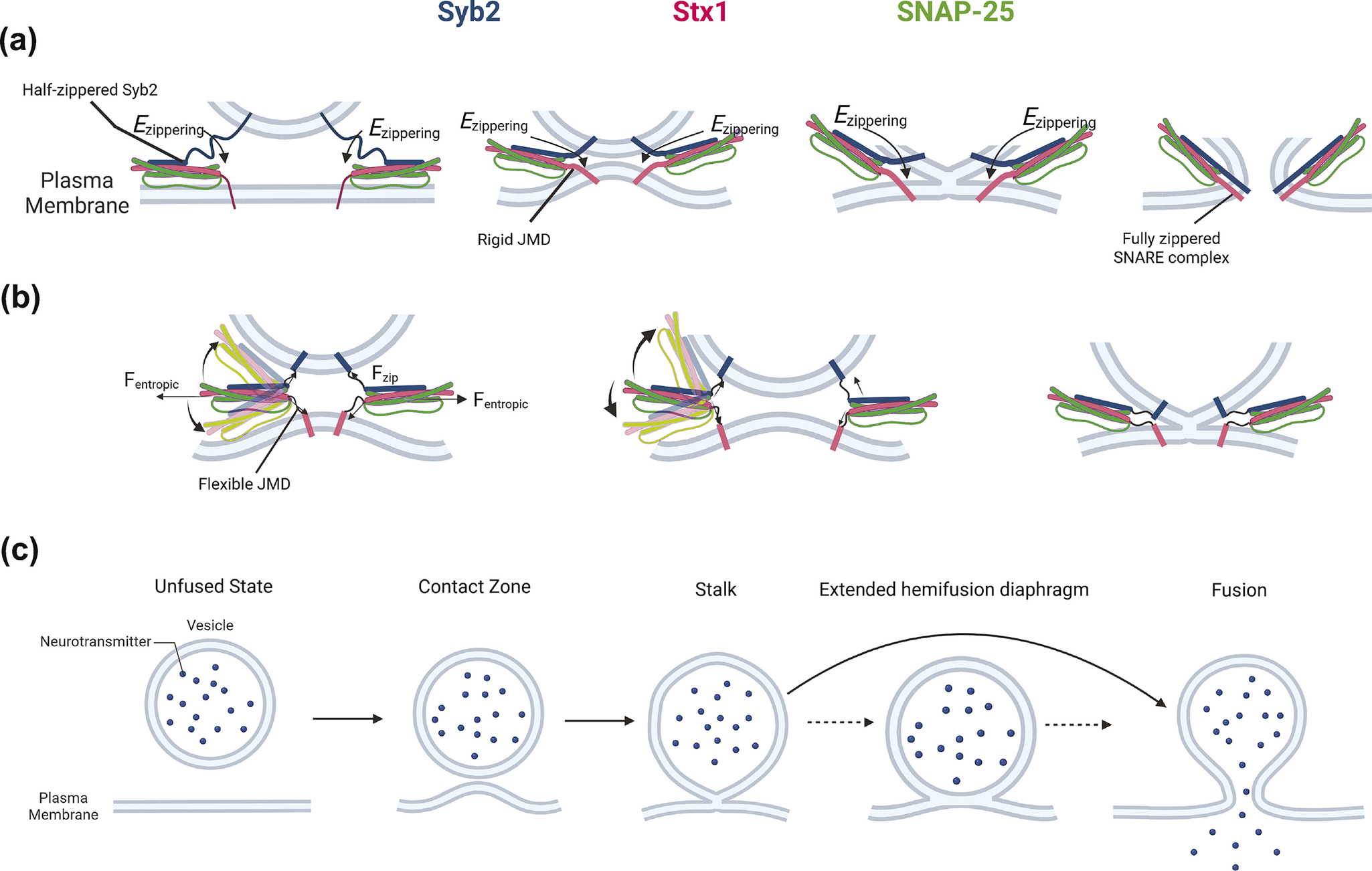
Figure 2. Contrasting hypotheses and shared features of how SNARE complexes drive membrane fusion.
(a) In the zippering-driven model, energy from SNARE complex zippering is transmitted via rigid and helical JMDs – represented by thick lines – which press the two membranes together, initiating stalk formation and subsequently FP opening. This process is facilitated by interactions between the C-terminal residues of Syb2 and Stx1 (Sharma and Lindau, Reference Sharma and Lindau2018; An et al., 2025). (b) In the entropic force model, thermal fluctuations of the SNDs generate entropic forces that expand the accessible conformational space for SND occupancy. In this case, zippering energy is dissipated, and flexible JMDs – represented by thin, curved lines – enable membrane contact, leading to reversible stalk formation and eventual FP opening (Butu et al., Reference Butu, An and O’Shaughnessy2025). (c) A generalized membrane fusion pathway shared by all MD simulations. The two membranes first adhere through a contact zone, followed by stalk formation via fusion of the cytoplasmic leaflet of the vesicle and the intracellular leaflet of the plasma membrane. The stalk then either expands into an extended HD – which can impede rapid fusion – or progresses to full fusion via FP formation.
The rigid JMD model aligns with the crystal structure of the post-fusion SNARE complex, which reveals a helical JMD (PDB: 3HD7) (Stein et al., Reference Stein2009). In 2011, Risselada et al. developed the first MARTINI-based membrane fusion system, in which two 20 nm vesicles were bridged by two SNARE complexes with structured and rigid JMDs (Risselada et al., Reference Risselada2011). Upon SNARE zippering, the SNARE complexes brought the membranes into close proximity, leading to FP formation within microseconds. However, when the JMDs were modeled as unstructured and flexible, fusion was abolished on this timescale. Similar results were later obtained in a vesicle–planar membrane fusion system, further reinforcing the importance of JMD rigidity (Risselada and Grubmuller, Reference Risselada and Grubmuller2012). This view is also supported by the CGMD simulations in ref. Fortoul et al. (Reference Fortoul2018) using a different force field.
Additional support for the rigid JMD hypothesis comes from simulations by Sharma and Lindau, who modeled fusion between a 40 nm vesicle and a planar plasma membrane bridged by six SNARE complexes (Sharma and Lindau, Reference Sharma and Lindau2018). Here, rigid JMDs and constrained distances to the center of mass of the layer 0 backbone beads facilitated spontaneous fusion within microseconds (Figure 2a). Moreover, AA simulations initiated from SNARE complexes with unstructured JMDs have demonstrated that progressive structuring and zippering of the SNARE JMDs enables lipid tail interdigitation, ultimately accomplishing FP formation on a similar timescale (Rizo et al., Reference Rizo2024).
However, this model has been challenged by findings that elongating the SNARE JMDs does not abolish fusion but instead slows down fusion kinetics (McNew et al., Reference McNew1999; Kesavan et al., Reference Kesavan2007). These findings led to the alternative hypothesis that SNARE-mediated fusion may primarily be driven by entropic forces rather than direct mechanical transduction (Mostafavi et al., Reference Mostafavi2017; McDargh et al., Reference McDargh2018; Butu et al., Reference Butu, An and O’Shaughnessy2025). To test this hypothesis, O’Shaughnessy’s group developed an UCG model capable of reaching millisecond timescales. In their early models, SNARE complexes were represented as rod-like chains of beads for the SNARE domain, with unstructured JMDs modeled using a worm-like chain representation, while membranes were treated as rigid surfaces (Mostafavi et al., Reference Mostafavi2017; McDargh et al., Reference McDargh2018). In a more recent iteration, TMDs were explicitly included, and lipid representations were adapted from previous studies (Cooke et al., Reference Cooke2005; Illya and Deserno, Reference Illya and Deserno2008) to allow direct visualization of membrane fusion rather than relying on inferred membrane-pressing energy (Butu et al., Reference Butu, An and O’Shaughnessy2025) (Figure 2b).
These simulations demonstrated that SNARE complexes could spontaneously generate ~8 pN of entropic force per complex, resulting in ~19 pN of membrane-pressing force per SNARE because of constant collisions and the trans-SNARE complex geometry which the vector of JMDs is bent ~67° away from that of SNARE domains in simulations. This force was sufficient to drive the formation of an extended fusion site, initiating membrane contact and ultimately leading to fusion (Butu et al., Reference Butu, An and O’Shaughnessy2025). Notably, these findings suggest that the presence of more SNARE complexes accelerates fusion kinetics.
Role of the JMD
The role of the SNARE JMD remains controversial, as evidence supporting both hypotheses has been reported. An early AA simulation study in 2003 found that increasing the concentration of the anionic lipid phosphatidylserine (PS) enhanced the helicity and rigidity of the Stx1A JMD (Knecht and Grubmuller, Reference Knecht and Grubmuller2003). CG simulations in MARTINI force field demonstrated that phosphatidylinositol 4,5-bisphosphate (PIP₂), a multivalent anionic lipid in the cytoplasmic leaflet of the plasma membrane, was enriched around the cationic Stx1A JMD (Sharma and Lindau, Reference Sharma and Lindau2017). Taken together, these studies suggest that the PIP₂ enrichment at the Stx1A JMD could further stabilize its helical structure and potentially facilitate transduction of the 30–85 kT zippering energy via the helical JMDs to squeeze together and fuse the two membranes. However, it is still unknown how much energy is transduced to nucleate membrane fusion because it is not clear how specific structural features are connected to the fusion pathways, and the involved energies are model-dependent (Jahn et al., Reference Jahn2024). Further, electron paramagnetic resonance (EPR) and nuclear magnetic resonance (NMR) experiments showed that the Stx1A JMD remained unstructured in vitro (Kim et al., Reference Kim2002; Lakomek et al., Reference Lakomek2019), challenging the notion that its helicity is a defining feature under all conditions. The structural characterization of these studies was, however, performed on isolated proteins and not on full SNARE complexes.
Pathway to fusion: From membrane contact to FP formation
A key question in membrane fusion research was whether the FP is proteinaceous or lipidic (Almers and Tse, Reference Almers and Tse1990; Zimmerberg et al., Reference Zimmerberg1993; Lindau and Almers, Reference Lindau and Almers1995; Jackson, Reference Jackson2010). Before the discovery of SNARE proteins, membrane fusion pathways had been studied for ~50 years, beginning with in vitro experiments showing that membrane fusion was induced solely by Ca2+ (Ginsberg, Reference Ginsberg1978; Ohki, Reference Ohki1982; Ohki and Ohshima, Reference Ohki and Ohshima1985; Rand et al., Reference Rand1985; Kachar et al., Reference Kachar1986; Nikolaus et al., Reference Nikolaus2010). Also, the conductance of FP has been studied extensively (Breckenridge and Almers, Reference Breckenridge and Almers1987; Hartmann and Lindau, Reference Hartmann and Lindau1995; Lindau and Almers, Reference Lindau and Almers1995; Han et al., Reference Han2004; Gong et al., Reference Gong2007). Based on experimental evidence, the widely accepted model of membrane fusion follows a sequential pathway involving: (1) membrane docking and priming, forming a potential fusion site (Fortoul et al., Reference Fortoul2015; Witkowska et al., Reference Witkowska2021; An and Lindau, Reference An and Lindau2024), (2) stalk nucleation (Sharma and Lindau, Reference Sharma and Lindau2018), and (3) FP opening from the stalk state (Chernomordik and Kozlov, Reference Chernomordik and Kozlov2008; Kozlov et al., Reference Kozlov2010; Fang and Lindau, Reference Fang and Lindau2014; Jahn et al., Reference Jahn2024) (Figure 2c). This pathway appears to be evolutionarily conserved, as extended hemifusion diaphragms (HDs) have been observed in both in vitro and in vivo studies but do not lead to rapid fusion (Figure 2c) (Diao et al., Reference Diao2012; Hernandez et al., Reference Hernandez2012; Zhao et al., Reference Zhao2016).
MD simulations have consistently reproduced this pathway (Risselada et al., Reference Risselada2011; Risselada and Grubmuller, Reference Risselada and Grubmuller2012; Sharma and Lindau, Reference Sharma and Lindau2018; Butu et al., Reference Butu, An and O’Shaughnessy2025). Initially, membrane adhesion and fusion site formation occur through either mechanical transduction of SNARE domain zippering or entropic forces. Subsequently, fusion proceeds via stalk nucleation, during which the TMD of Syb2 begins to approach that of Stx1. Finally, FP formation is facilitated by the hydrophilic C-terminal residues of Syb2 and Stx1. This pathway is also valid under membranes with asymmetric compositions (Sharma and Lindau, Reference Sharma and Lindau2018) introduced from ref (Sharma et al., Reference Sharma2015). Even though in some cases, additional intermediate structures, such as inverted micelles, have been occasionally observed (Risselada and Grubmuller, Reference Risselada and Grubmuller2012; Sharma and Lindau, Reference Sharma and Lindau2018), the overall membrane fusion pathway is similar among all MD simulations among different CG scales.
On the other hand, estimating the energy barriers between membrane docking, priming, stalk, hemifusion, and FP formation and the kinetics of the intermediate states based on MD simulations provides highly variable values among different force fields. Also, while extended HDs have been observed in multiple experimental studies (Nikolaus et al., Reference Nikolaus2010; Diao et al., Reference Diao2012; Hernandez et al., Reference Hernandez2012; Zhao et al., Reference Zhao2016), their roles in FP formation remain unclear. Instead of serving as an obligatory intermediate for rapid fusion, extended HDs may actually impair FP formation.
In CG simulations with MARTINI force field of a 40-nm vesicle fusing with a planar plasma membrane via six SNARE complexes – without constraints on the center-of-mass distances of layer 0 backbone beads – an extended HD formed, but no FP opened within 2 μs (Sharma and Lindau, Reference Sharma and Lindau2018). This also aligns with in vivo experimental observations where extended HDs persisted for timescales ranging from 0.2 to 26 s in ~15% of fusion events, even exceeding experimental observation windows in ~30% of fusion events, whereas ~55% of the events showed fusion occurred immediately with no observation of extended HDs (Zhao et al., Reference Zhao2016).
In contrast, MD simulations that exhibited rapid fusion – within microseconds for AA and CG simulations with MARTINI force field (Risselada et al., Reference Risselada2011; Risselada and Grubmuller, Reference Risselada and Grubmuller2012; Sharma and Lindau, Reference Sharma and Lindau2018) and within milliseconds for UCG models (Butu et al., Reference Butu, An and O’Shaughnessy2025) – did not report the presence of extended HDs. This discrepancy may be explained by theoretical analysis suggesting that extended HDs drain membrane tension and stabilize the HDs, thereby preventing HD rupture and FP formation (Warner and O’Shaughnessy, Reference Warner and O’Shaughnessy2012a, Reference Warner and O’Shaughnessy2012b; Warner et al., Reference Warner2023).
SNARE TMDs in FP formation
As originally reported for fusion mediated by Influenza hemagglutinin (HA) lacking the TMD and anchored in membranes via a glycosylphosphatidylinositol (GPI) tail, the GPI-anchored HA mediated lipid mixing (hemifusion) but no full fusion (content mixing) (Kemble et al., Reference Kemble1994). Correspondingly, it was found that lipid-anchored Syb2 failed to support fusion (Chang et al., Reference Chang2016). Several studies, including both experiments and MD simulations, indicate that TMDs play a crucial role in facilitating fast membrane fusion (reviewed in Fang and Lindau, Reference Fang and Lindau2014).
To investigate the function of the Syb2 TMD, Lindau et al. performed MARTINI MD simulations and found that ~70 kJ/mol of energy is required to extract Syb2 TMD from the synaptic vesicle (SV) membrane. This anchoring is stabilized by hydrophilic C-terminal residues as well as W89 and W90 (Lindau et al., Reference Lindau2012). Beyond its anchoring role, the Syb2 TMD has been shown to actively contribute to FP dynamics. Combining in vivo experiments with AA simulations, Dhara et al. demonstrated that Syb2 TMD flexibility catalyzes both FP formation and expansion (Dhara et al., Reference Dhara2016), and Han et al. showed that the flexibility of Syb2 TMD favors the helicity of JMD (Han et al., Reference Han2016). Similarly, MARTINI MD simulations have revealed that Stx1A TMD lowers the free energy barrier for stalk formation, likely by inducing local membrane dimpling near the TMD insertion site on the plasma membrane (Smirnova et al., Reference Smirnova2019).
Beyond the intrinsic properties of TMDs, post-translational modifications such as palmitoylation also modulate SNARE function. in vivo experiments by Vardar et al. showed that palmitoylation of the Stx1A TMD enhances spontaneous neurotransmitter release but does not significantly alter Ca2+-evoked release (Vardar et al., Reference Vardar2022). To further investigate the effects of Stx1A TMD palmitoylation, we constructed a MARTINI MD model of palmitoylated Stx1A (residues 189–288). These simulations suggest that palmitoylation stabilizes a Stx1A TMD conformation, resembling its structure within the Stx1A–SNAP25 t-SNARE complex, potentially facilitating t-SNARE assembly (An et al., 2025). However, FP simulations revealed that Stx1A palmitoylation only disrupts interactions with Syb2 TMD, which may delay FP opening. Additionally, analysis of Stx1A palmitoyl chain tilt angles suggests that the palmitoyl chains exert mechanical constraints on the FP, reducing both peak conductance and conductance fluctuations (An et al., 2025).
Syt: The calcium sensor in fusion machinery
Multiple Syt interaction modes with SNARE complex
Since the discovery of Syt as a Ca2+ sensor, the precise mechanism by which it interacts with the SNARE complex to trigger membrane fusion remains under debate. Over the past decade, studies have identified multiple potential interaction interfaces between Syt and SNAREs. The Brunger group identified two distinct interfaces: the primary interface (Zhou et al., Reference Zhou, Lai, Bacaj, Zhao, Lyubimov, Uervirojnangkoorn, Zeldin, Brewster, Sauter, Cohen, Soltis, Alonso-Mori, Chollet, Lemke, Pfuetzner, Choi, Weis, Diao, Südhof and Brunger2015) and the tripartite interface (Zhou et al., Reference Zhou2017. Additionally, Brewer et al. reported a polybasic interface where Syt–SNARE interactions occur (Brewer et al., Reference Brewer2015).
Although these interfaces have been proposed as sites for Ca2+-dependent regulation, it remains unclear which interaction mode (or combination thereof) is responsible for Ca2+ sensing and fusion clamping and triggering. Here, we review recent evidence and examine Syt–SNARE interactions through multiscale MD simulations to provide insights into their functional roles.
Ca2+-dependent lipid binding
One question about the Ca2+-regulation of Syt is focused on how Ca2+ binding alters interactions between its C2 domains (Figure 1b) and lipid molecules. To investigate Syt Ca2+ binding, AA simulations produced Syt1 structures with two Ca2+ ions chelated at the binding pocket of each C2 domain (Bykhovskaia, Reference Bykhovskaia2015). The simulations revealed that Syt1 conformational flexibility drastically increased upon Ca2+ binding. The separation distance between the C2A and C2B domains increased, and interdomain rotations became more frequent. These changes in the Syt1 conformational state, help initiating the fusion process. More recent studies demonstrated that Ca2+-bound C2 domains are enriched in anionic lipids such as PS and PIP₂ around the Ca2+-binding loops (CBLs), leading to significant membrane penetration of these loops (Bykhovskaia, Reference Bykhovskaia2021). Similarly, Bender et al. combined AA simulations with mass spectrometry and confirmed these Ca2+-dependent C2 domain–lipid interactions, along with anionic lipid preference interactions (Bender et al., Reference Bender2024). While C2A prefers an orientation more perpendicular to the membranes, C2B attaches parallel to the membrane. Courtney et al., combining experimental and AA simulation studies, revealed that Syt7 C2B shows stronger membrane penetration than Syt1 C2B (Courtney et al., Reference Courtney2023). Mehta et al., combining CG simulations with MARTINI3 force field and experiments showed that the Syt1 lysine-rich JMD regulates Ca2+ binding via liquid–liquid phase separations (LLPS) (Mehta et al., Reference Mehta2024). They proposed that these interactions play a key role in facilitating Ca2+-evoked neurotransmitter release.
To mimic Ca2 binding in the MARTINI2 force field aimed at extending the simulation timescale, we introduced charge-flip mutations (D303K, D309K, D363K, D365K, and D371K) in the C2B domain of Syt1. These mutations introduce more charge than the two Ca2+ ions that are thought to bind C2B (Fernandez et al., Reference Fernandez2001) to exaggerate the effect of Ca2+ binding and since more than 2 Ca2+ ions may be accumulated in the presence of PIP2 (Schiavo et al., Reference Schiavo1996). Membrane self-assembly simulations using the method of (Sharma et al., Reference Sharma2015), revealed the burial of CBLs into the membrane (Figure 3, unpublished results). These CG simulations with MARTINI force field successfully reproduced Ca2+-dependent PIP₂ enrichment in real plasma membrane composition, aligning with previous experimental and computational findings (Bykhovskaia, Reference Bykhovskaia2021; Bender et al., Reference Bender2024).
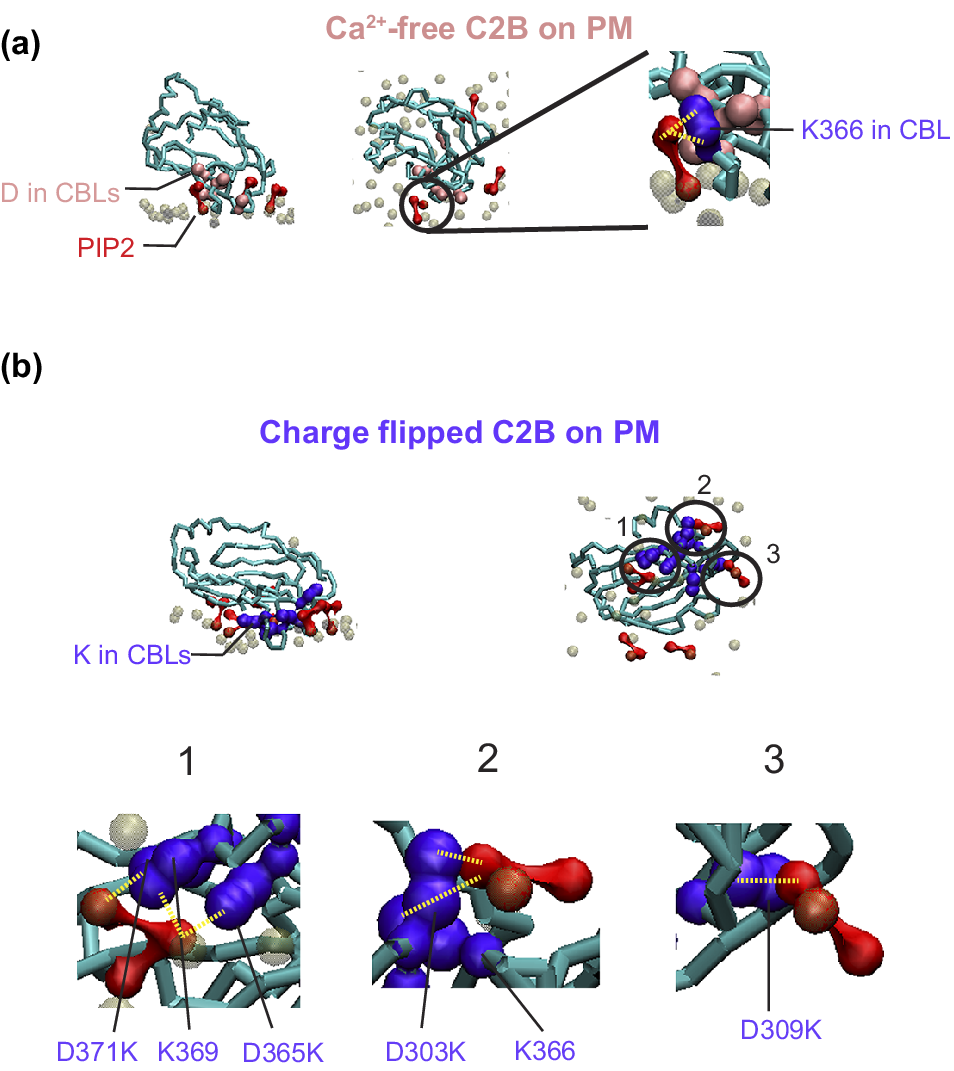
Figure 3. Charge-flip mutations in the C2B domain of Syt1 enhance interactions with PIP₂.
Ca2+-free (a) and charge-flip mutant (b) C2B domains of Syt1 (PDB: 5W5C (Zhou et al., Reference Zhou2017)) inserted into a plasma membrane via self-assembly, using the protocol described in Sharma et al. (Reference Sharma2015) with lipid composition from Sharma and Lindau (Reference Sharma and Lindau2018). Membranes were assembled in two stages: 50 ns simulations with x- and y-position restraints on the C2B domain, followed by 250 ns of unrestrained simulation. PIP₂ phosphate headgroup beads (PO4, P1 and P2) are colored red and the PO4 headgroup of the other phospholipids are colored brown transparently. The backbone of the C2B domains of Syt1 are colored in cyan. (a) Ca2+-free C2B domain: The left and middle panels show side and top views at 9 ns of unrestrained simulation, highlighting interactions between the calcium binding loops (CBLs) of the Ca2+-free C2B domain and PIP₂ headgroups. Anionic residues D303, D309, D363, D365, and D371 are shown in pink. Only one PIP₂ molecule interacts with the C2B domain in the circled region. The right panel zooms into this interaction, showing that the two phosphate groups on the inositol ring form a contact (~0.5 nm) with the sidechain of K366. (b) Charge-flip C2B mutant: The top panels show side and bottom views at 9 ns of unrestrained simulation for the charge-flip quintuple mutant D303K, D309K, D363K, D365K, and D371K (K; shown in purple). Three PIP₂ molecules are observed contacting the CBLs in the circled regions, driven by PIP₂–lysine interactions. The bottom row shows zoomed-in views of the three interaction regions. Region 1 shows one PIP₂ interacting with three lysine residues; region 2 with two lysines; and region 3 with one lysine – all at ~0.5 nm contact distance. The interacted lysines are annotated. This PIP₂ enrichment driven by the charge-flip mutant is consistent with prior all-atom simulations (Bykhovskaia, Reference Bykhovskaia2021; Bender et al., Reference Bender2024).
Ca2+-dependent effect on SNARE-mediated fusion
A key question in the study of Syt is how Ca2+-dependent lipid binding could alter SNARE conformation and dynamics to regulate FP formation. The primary interface has been proposed as the main site for transducing this effect, as its formation is independent of crystallization conditions (Zhou et al., Reference Zhou, Lai, Bacaj, Zhao, Lyubimov, Uervirojnangkoorn, Zeldin, Brewster, Sauter, Cohen, Soltis, Alonso-Mori, Chollet, Lemke, Pfuetzner, Choi, Weis, Diao, Südhof and Brunger2015, Reference Zhou2017), and its stability has been confirmed by AA simulations (Brunger and Leitz, Reference Brunger and Leitz2023).
Using a combination of AA simulations and NMR studies, the Rizo/Rosenmund groups identified Syt1 C2B domain interactions with SNAP-25 at two distinct regions within the SNARE complex: region I (including E295 and Y338) and region II (comprising R281, R398, and R399) (Figure 4a i) (Toulmé et al., Reference Toulmé2024; Jaczynska et al., Reference Jaczynska2025). Mutating E295 to alanine (E295A) enhanced the interactions at region I, leading to suppression of both Ca2+-evoked and spontaneous release, while interactions at region II remained intact (Toulmé et al., Reference Toulmé2024). Based on these findings, they proposed that C2B Ca2+ binding selectively modulates region I interactions, but not region II, to regulate Ca2+-triggered membrane fusion. Recent AA simulations by Rizo et al. found that Syt1 C2 domains hinder SNARE-mediated fusion if placed close to the fusion site and suggested that binding of Syt1 at the primary interface keeps it away from the fusion site such that it does not interfere with FP formation (Rizo et al., Reference Rizo, Jaczynska and Rosenmund2025). These studies led to the conclusion that Syt1 Ca2+ binding does not facilitate fusion by bridging membranes, inducing curvature, or perturbing membranes. They instead support the view Syt1 clamps fusion, being in the way at the fusion site and that Ca2+ binding may allow for its dissociation, enabling SNARE complex zippering. MARTINI (Sharma and Lindau, Reference Sharma and Lindau2018) as well as AA simulations (Rizo et al., Reference Rizo2024) have shown that in the absence of Syt, SNARE complex JMD linker zippering perturbs the bilayers, leading to FP formation.
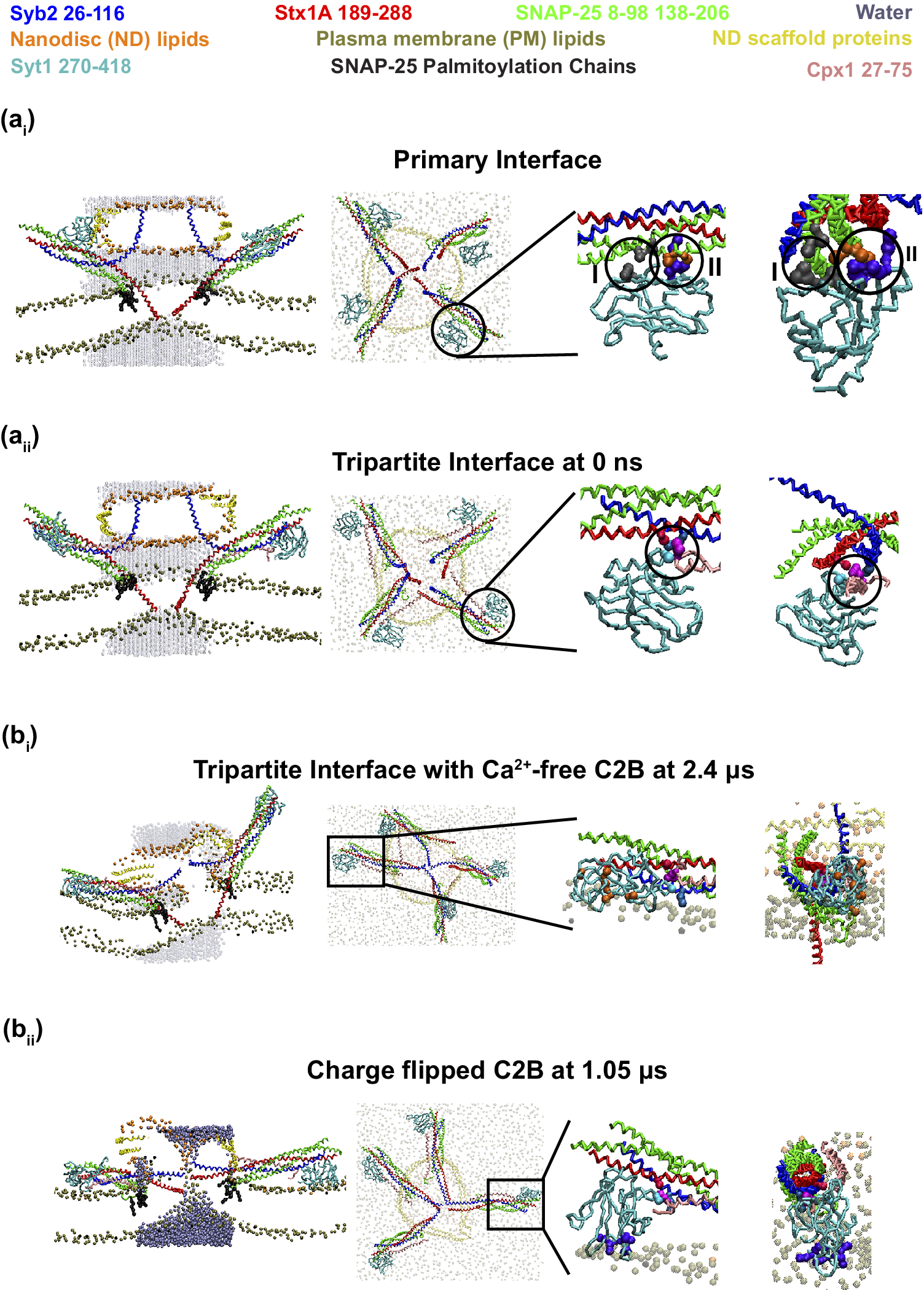
Figure 4. Regulation of Syt and Cpx in Neurotransmitter Release.
Syb2, Stx1A, SNAP-25, Syt1 C2B, and Cpx1 are colored blue, red, green, cyan, and pink, respectively. Water beads appear as transparent dark cyan, with one opaque bead marking an open FP. Lipid headgroups from the ND and plasma membrane are shown in orange and brown; the ND scaffold protein is yellow. SNAP-25 palmitoyl chains (black) are visible only in side views. (a) Initial configurations for FP simulations. Syt1 C2B domains were placed at the primary interface (a i) or tripartite interface with Cpx1 (a ii), by aligning crystal structure PDB 5W5C to the CG SNARE complex model (Sharma and Lindau, Reference Sharma and Lindau2018) using PyMOL (DeLano, Reference DeLano2002). Side and top views are shown, with transparent membranes and ND. Zoomed-in panels (right) display interface details from views perpendicular or parallel to the SNARE axis. Primary interface (a i): SNAP-25 interacts with C2B via Region I (SNAP-25: D166, E170; C2B: E295, Y338 colored in gray) and Region II (SNAP-25: D51, E52, E55 colored in orange; C2B: R281, R398, R399 colored in purple) circled in the two right panels. Tripartite interface (a ii): Syb2, Stx1A, C2B domain of Syt1 and Cpx1 interact together (Syb2: R47 (blue); Stx1A: E211 (red); C2B: T383 (cyan); Cpx1: Y70 (light purple)) at the circle region. (b) Ca2+-dependent effects on FP formation. (b i) With Ca2+-free C2B, 5/10 simulations stalled at hemifusion. At 2.40 μs, side/top views show incomplete SNARE zippering and Cpx1 AH crossing neighboring SNAREs as in Kümmel et al. (Reference Kümmel2011). Zoomed-in views show that CBLs remain unburied due to repulsive D residues (orange), though interprotein contacts shown above persist. (b ii) With charge-flip C2B, all 10 simulations formed FPs (mean lag: 779 ± 176 ns). At 1.05 μs, SNAREs are fully zippered, and an FP is visible. The Cpx1 AH aligns parallel to Syb2, and charge-flipped CBLs contact the PO₄ plane, consistent with Ca2+-triggered fusion. The Ca2+-dependent states of SNARE–Syt–Cpx complexes are consistent with Figure 6B of Kümmel et al. (Reference Kümmel2011).
Further doubts regarding the primary interface arose also from recent AA simulations, which suggested that Syt1 binding at this site forms a dead-end state, wherein Syt1 remains tightly attached to the plasma membrane but fails to insert into it upon Ca2+ binding (Bykhovskaia, Reference Bykhovskaia2024). Our own preliminary MARTINI FP simulations, in which a 10-nm nanodisc (ND) and a plasma membrane were bridged by four SNARE complexes unzipped up to layer 5 with C2B domains bound at each of the primary interfaces (Figure 4a i, unpublished results), showed no significant change in the lag time preceding the first appearance of an FP conductance when the Ca2+-bound mimic charge-flip mutations mentioned above were used (1452 ± 473 ns from 5 out of 10 simulations opened an FP within 4 μs for normal C2B versus 1512 ± 209 ns from 5 out of 10 simulations opened an FP within 4 μs for charge-flipped C2B, p = 0.917 in Kruskal test). The lag times were drastically increased compared to FP simulation without C2B bound at the primary interface (276 ± 20 ns from 10 out of 10 simulations opened an FP within 4 μs without C2B in ref. (An et al., 2025)). Moreover, the primary interface does not include Syt–Cpx interactions, which have been implicated in Ca2+-triggered fusion modulation (Bykhovskaia, Reference Bykhovskaia2024).
In contrast, our simulations suggest that Syt-SNARE binding at the tripartite interface, which includes Syt–Cpx interactions, facilitates FP opening in response to Ca2+ sensing – especially in the presence of the accessory helix (AH) of Cpx (Figure 4a ii, b, unpublished results). This aligns with experimental findings showing that tripartite interface interactions are highly dynamic (Jaczynska et al., Reference Jaczynska2023), increasing the likelihood that Ca2+-dependent C2 domain–lipid interactions may disrupt Syt1–SNARE interactions to enable fusion.
Cpx: The fusion clamp and facilitator
Dual role of complexin in spontaneous and Ca2+-evoked fusion
Cpx plays a paradoxical role in membrane fusion. Experimental evidence suggests that its N-terminal domain (NTD) acts as a catalyst for Ca2+-evoked fusion, while its C-terminal domain (CTD) functions as an inhibitor of spontaneous fusion (Figure 1b) (Martin et al., Reference Martin2011; Jorquera et al., Reference Jorquera2012; Li et al., Reference Li2024).
Despite this functional dichotomy, crystal structures reveal that the central helix (CH) of Cpx forms stable interactions with the groove between Syb2 and Stx1A within the SNARE complex (Bracher et al., Reference Bracher2002; Chen et al., Reference Chen2002; Kümmel et al., Reference Kümmel2011), even in the presence of the Syt1 C2B domain at the tripartite interface (Zhou et al., Reference Zhou2017. This raises a fundamental question: How does Syt1 regulate SNARE complex conformation and dynamics in a Ca2+-dependent manner? To address this question, we review recent MD simulations of SNARE-mediated fusion that incorporate both the AH and CH of Cpx, shedding light on its regulatory mechanisms in SNARE-mediated fusion.
MD insights into Cpx inhibitory function
MD simulations of Cpx–SNARE interactions began in 2010 with Ghahremanpour et al., who investigated the role of the CH and AH of Cpx in SNARE complex interactions (Ghahremanpour et al., Reference Ghahremanpour2010). Their AA simulations revealed: (1) The CH forms salt bridges and hydrogen bonds with the SNARE complex. (2) Cpx can also bind the t-SNARE complex (Stx1A and SNAP-25) in the absence of Syb2, reducing t-SNARE flexibility. (3) The AH and the C-terminal region of Syb2’s SNARE domain interact with Stx1A, stabilizing the complex. However, AH proximity to Syb2 destabilizes Stx1A and the SN1 helix of SNAP-25, suggesting that Cpx inhibits membrane fusion by competing with Syb2 for t-SNARE binding.
In 2013, Bykhovskaia et al. combined AA simulations and in vivo experiments, confirming that Cpx interacts with Syb2 and SNAP-25 via salt bridges (Bykhovskaia et al., Reference Bykhovskaia2013). These interactions stabilize SNARE complex unzippering up to layer 7, modulating the fusion process. Further studies demonstrated that Cpx–Syb2 interactions increase Cpx flexibility, preventing Syb2 from fully zippering and thereby clamping spontaneous fusion (Vasin et al., Reference Vasin2016; Brady et al., Reference Brady2021).
Recently, Rizo et al. conducted AA simulations incorporating both the AH and CH of Cpx1 into a system containing a 20 nm vesicle and a planar membrane bridged by four SNARE complexes (Rizo et al., Reference Rizo2022). Their findings showed that: (1) CH–SNARE interactions (via Y70) remained stable throughout the entire simulation. (2) The AH primarily interacted with the synaptic vesicle, likely exerting a steric effect that prevents membrane contact and fusion initiation. However, this steric hindrance may require longer simulation timescales to be fully explored.
Effects of Syt1 on Cpx-mediated SNARE regulation
Syt1 interacts with SNARE complexes not only at the primary interface but also at the tripartite interface, which is less stable and also involves Cpx (Zhou et al., Reference Zhou2017). Given the complementary roles of Syt and Cpx, this interface is a potential candidate to regulate the transition enabling FP formation. We therefore performed preliminary MARTINI FP simulations of the tripartite interface incorporating Syt1, AH, and CH of Cpx1 in addition to the SNARE complexes. We found that the FP formation dynamics mediated by SNARE complexes was highly dependent on whether charge-flip mutations were applied to the C2B domain of Syt1: (1) In simulations with wild-type C2B, only 5 out of 10 trials resulted in FP formation within 4 μs. The mean lag time was 1278 ± 270 ns calculated from these five simulations. (2) In contrast, in simulations with charge-flipped C2B, 10 out of 10 trials resulted in FP formation, and the lag time was 779 ± 176 ns, even if p = 0.066.
Additionally, in wild-type C2B simulations, the AH of Cpx frequently interacted with the cytoplasmic leaflet of the ND and the intracellular leaflet of the plasma membrane during the stalk state, preventing full SNARE zippering (Figure 4b i, unpublished results). SNARE complex zippering was occasionally obstructed by the AH attached to the neighbour SNARE complex. However, with charge-flipped C2B, SNARE complexes adopted orientations that facilitated the AH of Cpx staying parallel to the zippered SNARE complex, thereby enabling full zippering and FP opening (Figure 4b ii, unpublished results). These observations seem consistent with the proposal that FP opening is enabled when the zippering obstruction by Cpx AH is removed (Kümmel et al., Reference Kümmel2011). Furthermore, combining AA simulations with in vitro experiments, Courtney et al., discovered that the Cpx C-terminal amphipathic helix stabilizes the FP via membrane interactions (Courtney et al., Reference Courtney, Wu, Mandal, Swift, Zhang, Alaghemandi, Wu, Bradberry, Deo, Lavis, Volkmann, Hanein, Cui, Bao and Chapman2022).
Back-mapping CG simulations to AA models: A case study on FP conductance
CG molecular representations are widely applied in MD to extend simulation timescales by reducing the atomic details of molecular systems. In some ultra-coarse-grained models, solvent molecules are represented implicitly, allowing simulations to reach millisecond timescales. However, this simplification comes at the cost of reduced accuracy in molecular interactions due to the loss of atomic resolution.
To overcome this limitation back-mapping of CG simulations to AA models has emerged as a powerful approach. CG simulations are employed to explore long-timescale phenomena and sample conformational space efficiently and then use back-mapping to refine the results and obtain high-resolution insights into specific interactions or structural features. This strategy allows for a more comprehensive understanding of complex biological processes, balancing the need for computational efficiency with the desire for atomic-level accuracy. Here, we review studies integrating MARTINI coarse-grained and AA force fields alongside experiments to uncover the molecular determinants of FP conductance (Sharma and Lindau, Reference Sharma and Lindau2018; Delacruz et al., Reference Delacruz2021).
In these studies, the FP configuration was initially generated using CG simulations with MARTINI force field, as the computational simulation time required for FP opening is extremely long for direct AA simulations. The MARTINI force field represents four non-hydrogen atoms as a single coarse-grained bead, and groups four water molecules into a single water bead. This reduces thermal fluctuations, smooths the potential energy surface, and allows for longer simulation time steps, making millisecond-scale simulations feasible.
While CG simulations with MARTINI force field effectively capture bulk properties of proteins, membranes, and water, they can underestimate molecular interactions in nanometer-sized constricted pores, affecting the quantification of water and ion diffusivities. To address this, the system was back-mapped to an atomistic representation to recover finer molecular details. Back-mapping revealed a critical conductance threshold (~300 pS), below which ion selectivity emerged: cationic neurotransmitters (e.g., dopamine) were preferentially selected and permeable, whereas anionic neurotransmitters (e.g., glutamate) were blocked due to the anionic microenvironment within the narrow FP (Delacruz et al., Reference Delacruz2021). These findings highlight the necessity of back-mapping to accurately capture molecular interactions that govern neurotransmitter permeation and synaptic signaling. In addition, complementarity between AA and CG simulations was used to parametrize CG models and quantify the SNARE complex dynamics including the change of protein secondary structures (Darre et al., Reference Darre2012; Zheng, Reference Zheng2014).
Limitations and perspective
Limitations of current simulations
MD simulations are powerful tools for studying membrane fusion, but they are inherently limited by factors such as force field accuracy, system size, simulation timescale, and incomplete structural knowledge of proteins and complexes.
First, limitations in force fields – particularly in CG models – alongside constraints in system size and accessible timescales, cannot be simultaneously resolved without substantially increased computational resources. To simulate larger systems over longer timescales without increasing computational demand, more coarse-grained force fields must be used, at the cost of reduced chemical interaction resolution. As a result, dynamic properties like lipid diffusion are accelerated in CG models (Marrink et al., Reference Marrink2004; Ingolfsson et al., Reference Ingolfsson2017). Furthermore, compromising chemical interaction resolution by CG can alter the energy landscape of interactions among components of the fusion machinery and that of interactions between the protein fusion machinery and lipids. The energy barriers of protein dynamics and membrane remodeling ultimately affect the kinetics of FP formation.
System size limitations can also introduce artifacts. When the natural diffusive distance of molecules exceeds the simulation box dimensions, periodic boundary conditions may induce non-physical dynamics or artificial interactions. To fit within these limitations, artificial geometries – such as 20 nm vesicles or 10 nm NDs – are often used instead of physiologically relevant 50 nm synaptic vesicles, which may compromise biological realism.
Likewise, timescale limitations pose a major challenge. Physiological fusion occurs on submillisecond timescales, which are difficult to reach, even with CG simulations. To observe fusion events within accessible timescales, researchers often introduce dynamics-accelerating factors, such as fusogenic lipid compositions or pre-constrained SNARE complex structures, based on the selected force field.
Beyond these intrinsic limitations, external limitations also exist – most notably, the lack of precise structural information for several key components of the fusion machinery. Although in vitro structures of post-fusion SNARE complexes and SNARE–Syt–Cpx assemblies have been resolved and are often used to derive initial conditions for simulations, the native, dynamic assemblies that govern membrane docking, priming, and fusion remain elusive due to their inherent structural flexibility and transient nature.
For example, Munc13 and Munc18 remain difficult to incorporate into simulations because their interaction interfaces with the SNARE complex and membranes are still unresolved. As a result, it is unclear how synaptic vesicles dock to the plasma membrane and how Syb2, Stx1, SNAP-25, Syt, and Cpx assemble into functional pre-fusion complexes with assistance from Munc13 and Munc18. Without knowledge of these 3D pre-fusion structures or the molecular architecture of vesicle docking sites, the physiologically accurate initial conditions for simulating neurotransmitter release remain uncertain. This in turn obscures our understanding of the temporal distribution of vesicle docking, priming, and fusion events during exocytosis.
To address this gap, Li et al used cryo-electron tomography to observe a symmetrical arrangement of six core components – likely including SNAREs, Syt, Cpx, and Munc13 – at the fusion site in nerve growth factor-differentiated neuroendocrine PC12 cells (Li et al., Reference Li2019). Similar structural features were later observed in mouse hippocampal neurons (Radhakrishnan et al., Reference Radhakrishnan2021). Building on this, the Rothman group proposed – based on reconstitution experiments in (Bera et al., Reference Bera2023) – that 12 SNARE complexes may assemble in a two-step process, coordinated by associated proteins such as Syt, Cpx, Munc13, and synaptophysin, forming a highly symmetric, ring-like structure (Rothman et al., Reference Rothman2023). However, the detailed 3D structures of these assemblies are yet to be resolved.
Because of these challenges, using accurate protein complexes in fusion simulations is essential for generating realistic, dynamic representations of membrane fusion. Computational resources that reliably predict protein folding and protein–protein interactions are critical for addressing the current uncertainties surrounding physiological initial conditions.
In the following subsection, we briefly discuss structure prediction tools such as AlphaFold (https://deepmind.google/science/alphafold/), Robetta (https://robetta.bakerlab.org/), and SWISS-MODEL (https://swissmodel.expasy.org), which are capable of predicting the 3D structures of individual proteins and protein complexes. These tools are particularly valuable in reconstructing unknown assemblies involved in membrane fusion – especially following the development of AlphaFold2, which has significantly improved the accuracy of structure predictions (Bertoline et al., Reference Bertoline2023).
Completing protein structures and assembling complex systems
Filling the gaps between protein sequences and their corresponding 3D structures has been extensively studied for ~30 years. M. C. Peitsch established the first free server called SWISS-MODEL (Peitsch, Reference Peitsch1996) to predict the protein 3D structures based on the homologous protein structures (Waterhouse et al., Reference Waterhouse2018). This method is called Homology, which stems from the observation that the interacting interfaces among homologous complexes are often conserved (Zhang et al., Reference Zhang2010), with the availability of conserved protein–protein interaction templates (Kundrotas et al., Reference Kundrotas2012). In 2018, SWISS-MODEL was able to model both homomeric and heteromeric complexes (Waterhouse et al., Reference Waterhouse2018). Another server called Robetta was established in Baker’s lab applying Rosetta-based algorithms for de novo structure prediction (Rohl et al., Reference Rohl2004). In the Rosetta method, short protein fragments are assembled in Monte Carlo strategy to generate native-like 3D conformations. This method yields very accurate predictions (0.3–0.6 nm alpha-carbon RMSD) compared to the experimental structures for proteins with 60 or more residues.
In the 2020s, with the increasing number of experimental solved structures (~ 240 k PDB structures archived in https://www.rcsb.org/) and the maturation of deep learning models, AI has been introduced into predicting protein 3D structures based on their amino acid sequences. According to the 14th Critical Assessment of Structure Prediction (CASP14) conference (https://predictioncenter.org/casp14/zscores_final.cgi), Alphafold2 and Robetta (using RoseTTAFold network) servers are the top2 most accurate models to predict protein 3D structures (Baek et al., Reference Baek, DiMaio, Anishchenko, Dauparas, Ovchinnikov, Lee, Wang, Cong, Kinch, Schaeffer, Millán, Park, Adams, Glassman, DeGiovanni, Pereira, Rodrigues, van Dijk, Ebrecht, Opperman, Sagmeister, Buhlheller, Pavkov-Keller, Rathinaswamy, Dalwadi, Yip, Burke, Garcia, Grishin, Adams, Read and Baker2021; Bryant et al., Reference Bryant2022) and have been awarded the Nobel prize in Chemistry in 2024 (Callaway, Reference Callaway2024). In addition, ColabFold is derived from Alphafold2 by improving the sequence search, providing powerful tools for modeling homomer and heteromer complexes (Mirdita et al., Reference Mirdita2022). ColabFold also exposes advanced functionality, expands the environmental databases, and enables large protein structure prediction scale, at about a 90-fold speed-up over AlphaFold2. Furthermore, Harmalkar et al., developed the improved version of ReplicaDock called AlphaRED (AlphaFold-initiated Replica Exchange Docking) by combining AlphaFold as a structural template generator to accurately predict protein–protein docking (Harmalkar et al., Reference Harmalkar2025). Thus, AI models like AlphaFold (Jumper et al., Reference Jumper2021), Colabfold (Mirdita et al., Reference Mirdita2022) and Robetta (Baek et al., Reference Baek, DiMaio, Anishchenko, Dauparas, Ovchinnikov, Lee, Wang, Cong, Kinch, Schaeffer, Millán, Park, Adams, Glassman, DeGiovanni, Pereira, Rodrigues, van Dijk, Ebrecht, Opperman, Sagmeister, Buhlheller, Pavkov-Keller, Rathinaswamy, Dalwadi, Yip, Burke, Garcia, Grishin, Adams, Read and Baker2021) can help to address some of these external limitations by providing structural predictions for components of the fusion machinery. For more details, please see the minireview of Bertoline et al. (Reference Bertoline2023).
This advancement is particularly valuable for studying synaptic fusion machinery, as it allows for the modeling of full-length SNARE proteins and accessory factors such as Syt, Cpx, Munc13, and Munc18 to further approach a realistic structure of the fusion machine mediating transmitter release.
For instance, by aligning AlphaFold-predicted structures of Syb2, Stx1A, and SNAP-25 with crystal structures of the SNARE complex (PDB: 3HD7) (Stein et al., Reference Stein2009), a near-complete model of the synaptic vesicle fusion system can be assembled (An and Lindau, Reference An and Lindau2024). Similarly, tripartite interfaces involving Syt and Cpx can be reconstructed by integrating AlphaFold-predicted structures with known crystal structures of their interactions (PDB: 5W5C) (Zhou et al., Reference Zhou2017).
Exploring isoform-specific differences
Beyond completing incomplete protein structures, AI models also facilitate the modeling of isoform-specific variations, offering insights into functional differences between isoforms. By aligning AlphaFold-predicted isoform structures and replacing the canonical protein with its isoform in MD simulations, it has been possible to investigate how sequence variations strengthen or weaken key interactions, ultimately influencing synaptic vesicle fusion.
For example, increased fusogenicity altering the size of the primed vesicle pool and the clamping of spontaneous neurotransmitter release of Stx2 compared to Stx1A has been attributed to sequence differences in the C-terminal Stx SNDs (Lázaro et al., Reference Lázaro2024). MD simulations could provide molecular insights into such Stx isoform differences, explaining why Stx2 SNARE complexes are more fusogenic than Stx1A SNARE complexes.
Similarly, MD simulations could be used to explore isoform-specific differences in accessory proteins. For example: (1) Syt1 versus Syt7, elucidating their distinct roles in fusion and docking for synchronous versus asynchronous release (Vevea et al., Reference Vevea2021; Wu et al., Reference Wu2024), and how different membrane penetrations (Courtney et al., Reference Courtney2023) alter SNARE–Syt interactions to fine-tune FP kinetics and (2) Cpx1 versus Cpx2, illustrating different roles of Cpxs in neurotransmission like Cpx2 rather than Cpx1 facilitates large dense core vesicle docking and priming in chromaffin cell exocytosis (Cai et al., Reference Cai2008).
Integrating AlphaFold-predicted structures with MD simulations offers great promise to bridge structural and functional gaps, and lead to a more comprehensive understanding of how protein isoforms modulate neurotransmitter release dynamics. This approach could also further provide guidance on future in vitro and in vivo experiments like mutating key residues to observe phenotypes to deeply understand the mechanisms of neurotransmission and the related diseases due to neurotransmitter release dysfunctions.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this study the first author Dong An used ChatGPT in order to improve language and readability. After using this tool/service, the author(s) reviewed and edited the content as needed and took full responsibility for the content of the publication.
Acknowledgements
We thank Reinhard Jahn and Michael Kozlov for their critical reading of the manuscript and excellent comments and suggestions. Unpublished simulations were performed on Triton cluster from Frost Institute for Data Science and Computing (https://idsc.miami.edu/) and Bridges-2 at Pittsburgh Supercomputing Center through allocation BIO250019 and BIO250054 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296. Figures 1a and 2 were created with BioRender.com.
Financial support
This study has been supported by US National Institutes of Health (NIH) grant R35GM139608.
Competing interests
The authors declare none.





