In 2009, as part of the Genetic Association Information Network (GAIN) initiative, we published (Sullivan et al., Reference Sullivan, de Geus, Willemsen, James, Smit, Zandbelt and Penninx2009) the first genome-wide association study (GWAS) for major depressive disorder (MDD). The participants were assembled from two large Dutch cohorts: the Netherlands Study of Depression and Anxiety (NESDA) and the Netherlands Twin Registry (NTR). A comparison between 1,738 cases with lifetime diagnosis of DSM-IV MDD and 1,802 screened controls highlighted a signal overlapping the region of PCLO gene, although variants identified did not reach genome-wide significance. Importantly, our result received strong support (though not reaching formal significance) from Australian data similarly ascertained from a population twin registry (Sullivan et al., Reference Sullivan, de Geus, Willemsen, James, Smit, Zandbelt and Penninx2009). Based on the promising role of PCLO encoding a cytoskeletal protein located in the presynaptic active zone, the original finding was followed by replication efforts, with positive (Hek et al., Reference Hek, Mulder, Luijendijk, van Duijn, Hofman, Uitterlinden and Tiemeier2010) and negative (Verbeek et al., Reference Verbeek, Bevova, Bochdanovits, Rizzu, Bakker, Uithuisje and Heutink2013) results. In the present study, we revisited the original GWAS after introducing important improvements. The overall sample size was almost doubled, adding newly genotyped participants from the original cohorts, consisting mainly of a large number of controls carefully screened using several sources of information including longitudinal assessments and family history (Boomsma et al., Reference Boomsma, Willemsen, Sullivan, Heutink, Meijer, Sondervan and Penninx2008). The number of interrogated SNPs was increased by re-genotyping the majority of participants on a newer array and by imputing variants with a denser reference panel (1,000 Genomes Phase 3).
The present sample consisted of 6,507 participants (63.1% females) of European ancestry from NESDA and NTR. Lifetime MDD diagnoses according to DSM-IV were ascertained using the Composite Interview Diagnostic Instrument. Healthy controls were screened based on absence of any lifetime psychiatric disorder (NESDA), no report of MDD, no known first-degree relatives with MDD, and a low factor score based on a multivariate analysis of depressive complaints, anxiety, neuroticism, and somatic anxiety (Boomsma et al., Reference Boomsma, Beem, van den Berg, Dolan, Koopmans, Vink and Slagboom2000). After applying stringent quality control criteria (Supplementary Methods), we performed a GWAS comparing 1,942 cases and 4,565 controls, adjusting for gender and 10 ancestry-informative principal components. Family structure was taken into account using a sandwich estimator implemented in Plink.
The estimated LD score regression intercept was 1.03, suggesting no appreciable inflation of the test statistics attributable to unaccounted stratification (Figure S1). The analysis identified 17 genome-wide significant SNPs (p < 5 × 10−8; Figure 1) spanning the PCLO gene on chromosome 7 with rs2715157 emerging as the SNP most significantly associated with MDD (p = 2.91 × 10−8); its minor allele (A) was negatively associated with MDD (OR = 0.79). The PCLO lead SNP identified in the original GWAS, rs2715148 showed the same effect size as in the original analysis but was now genome-wide significantly associated with MDD (p = 3.89 × 10−8; OR = 0.79). Similarly, the non-synonymous coding SNP rs2522833 reported the same effect size of the original GWAS but a smaller p value (OR = 1.26, p = 8.29 × 10−8). These SNPs were highly correlated with rs2715157 (r 2 > 0.8; Figure 2). Furthermore, in silico functional annotation of the top SNPs showed possible functional effects. The PCLO SNP rs2715157 alters the sequence of six protein-binding motifs including Pou3f2, indicating a possible effect on the activation of corticotrophin releasing hormone. It also bound to the P300 protein known to regulate gene transcription. SNPs rs2751161 and rs2522835 were contained in a DNase I hypersensitive site suggesting open chromatin, although they did not alter any of the transcription factor binding sites present in the promoter flanking region. Together, these data indicate these SNPs might have direct functional roles (Table S1). Finally, p values of analyzed SNPs were used to perform a gene-based test (Supplementary Methods), and PCLO showed a significant association with MDD (p = 1.48 × 10−7), even considering a stringent multiple comparison adjustment accounting for 20,000 genes (p = 2.5 × 10−6).

FIGURE 1 A. Genome-wide association results for MDD in NTR-NESDA cohorts. The y axis denotes the -log10 (p value) for association. The x axis gives the physical position of SNPs across the genome. The red line indicates the threshold of genome-wide significance p < 5 × 10−8, and the blue line indicates the threshold of p < 5 × 10−5. B. Genome-wide significant loci in the GWAS for MDD in NTR-NESDA cohorts.
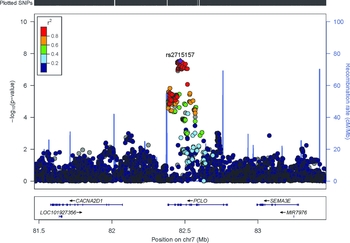
FIGURE 2 Regional association plot of PCLO region. The -log10 p values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The estimated recombination rates from the 1,000 Genomes Project March 2012 release are shown as blue lines, and the genomic locations of genes within the regions of interest in the NCBI Build 37 human assembly are shown as arrows. SNP color represents LD with the most highly associated SNP. The figure was created with LocusZoom (http://csg.sph.umich.edu/locuszoom/).
These findings provide evidence for an association between the PCLO gene and MDD that now clearly reaches genome-wide significance. An important factor was the increase in sample size. Indeed, the effect size detected for SNPs present in both the original and current analyses were exactly the same, but the improved precision in their estimation impacted the test significance. Allele frequencies were also the same for SNPs present in both analyses, suggesting no probable effect on results due to technical differences between the genotyping platforms used in the two GWASs. Nevertheless, imputation allowed us to examine additional SNPs in the same LD block previously not included.
It is important to note that the effectiveness of sample size increase may have been amplified by the clinical homogeneity of the sample and the detailed phenotyping procedure. Recently, the assembling of massive samples has enabled the success of GWAS for depression (Hyde et al., Reference Hyde, Nagle, Tian, Chen, Paciga, Wendland and Winslow2016; Okbay et al., Reference Okbay, Baselmans, De Neve, Turley, Nivard, Fontana and Cesarini2016), overcoming the problematic phenotypic heterogeneity of this trait. Nevertheless, large collaborative studies clearly identified substantial heterogeneity among different MDD datasets as indexed by cross-dataset SNP-correlations smaller than one (Lee et al., Reference Lee, Ripke, Neale, Faraone, Purcell, Perlis and Wray2013), possibly indicating the presence of dataset specific variants. Therefore, collecting a larger sample size, although on a smaller scale, in more homogeneous populations still constitutes a valuable complementary effort aimed at clarifying specific genetic signals that might be overshadowed by the heterogeneity arising from the addition of each new dataset in meta-analytic studies.
To conclude, in our current genome-wide study of MDD, we found confirming evidence of a genome-wide significant association with the PCLO gene. Our findings, together with previous biological evidence (Ahmed et al., Reference Ahmed, Chioza, Rajab, Schmitz-Abe, Al-Khayat, Al-Turki and Mochida2015; Waites et al., Reference Waites, Leal-Ortiz, Andlauer, Sigrist and Garner2011), suggest the importance of the PCLO gene to MDD, which is worth further replication and functional studies.
Conflict of Interest
None.
Acknowledgments
Netherland Twin Register and Netherlands Study of Depression and Anxiety (NESDA): Funding was obtained from the Netherlands Organization for Scientific Research (NWO) and MagW/ZonMW grants Middelgroot-911-09-032, Spinozapremie 56-464-14192, Geestkracht program of the Netherlands Organization for Health Research and Development (ZonMW 10-000-1002), Center for Medical Systems Biology (CSMB, NOW Genomics), Genetic influences on stability and change in psychopathology from childhood to young adulthood (ZonMW 912-10-020), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI–NL, 184.021.007), VU University's Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); the European Science Council (ERC Advanced, 230374). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health, Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA) and the National Institutes of Health (NIH R01 HD042157-01A1, MH081802, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995).
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2017.30




