MZ twin pairs result from the fertilization and subsequent splitting of a single embryo and, thus, are widely considered to be genetically identical. Being genetically identical makes MZ twins an important resource for the study of developmental plasticity in humans. A growing number of studies are investigating epigenetic differences in phenotypically discordant MZ twins, particularly those discordant for disease (Bell & Spector, Reference Bell and Spector2011; Kato et al., Reference Kato, Iwamoto, Kakiuchi, Kuratomi and Okazaki2005; Van Dongen et al., Reference Van Dongen, Slagboom, Draisma, Martin and Boomsma2012; Zwijnenburg et al., Reference Zwijnenburg, Meijers-Heijboer and Boomsma2010).
While MZ twins are largely genetically identical, somatic mutations have been observed in one twin of a pair. These tend to be observed in twin pairs that are discordant for disease, although it is likely that such differences also exist unnoticed within MZ pairs with ‘normal’ phenotypes. For example, mutations have been found in ATP2A2 in the affected twin of a pair discordant for Darier disease (Sakuntabhai et al., Reference Sakuntabhai, Ruiz-Perez, Carter, Jacobsen, Burge, Monk and Hovnanian1999), in IRF6 in a pair discordant for Van der Woude syndrome (Kondo et al., Reference Kondo, Schutte, Richardson, Bjork, Knight, Watanabe and Murray2002) and in SCN1A in Dravet's syndrome (Vadlamudi et al., Reference Vadlamudi, Dibbens, Lawrence, Iona, McMahon, Murrell and Berkovic2010).
Copy-number variants (CNV) are becoming recognized as affecting a wide range of human traits (Girirajan et al., Reference Girirajan, Campbell and Eichler2011; Zhang et al., Reference Zhang, Gu, Hurles and Lupski2009) and thus there is potential for MZ discordance to be caused by de novo CNVs. An initial investigation of 19 MZ twins pairs — 9 pairs discordant for disease and 10 phenotypically unselected or concordant normal pairs — found three copy-number differences within pairs, two large deletions in one individual of a discordant pair, and one large deletion in a member of a phenotypically concordant pair (Bruder et al., Reference Bruder, Piotrowski, Gijsbers, Andersson, Menzel, Sandgren and Dumanski2008). All these CNVs were present in only a portion of cells in the carrier, with frequencies of 20% and 15% for the CNV in the discordant pair and in 80% of cells in the individual from the concordant pair. A study of CNV in MZ twin pairs selected for concordance and discordance for attention problems identified two de novo CNVs, one occurring pre-twinning (and thus present in both members of the twin pairs) and one post-twinning (Ehli et al., Reference Ehli, Abdellaoui, Hu, Hottenga, Kattenberg, van Beijsterveldt and Davies2012). A number of other studies of MZ twins with discordant disease phenotypes have failed to identify any CNV discordance within the pair (Lasa et al., Reference Lasa, Ramón y Cajal, Llort, Suela, Cigudosa, Cornet and Baiget2010; Ono et al., Reference Ono, Imamura, Tasaki, Kurotaki, Ozawa, Yoshiura and Okazaki2010; Veenma et al., Reference Veenma, Brosens, de Jong, van de Ven, Meeussen, Cohen-Overbeek and de Klein2012).
A thorough quantification of the rate of genetic differences in a phenotypically unselected set of MZ twins is required in order to determine the population rate of copy-number differences within MZ pairs. If such copy-number differences are common within unselected MZ pairs, the probability of any observed copy-number differences in an MZ pair discordant for a disease being causal is greatly reduced. The frequency of copy-number differences is also an important consideration when investigation developmental plasticity in epigenetic studies of MZ twins as such studies assume genetic identity within the twin pair. We address this issue through the investigation of CNV in a population-based sample of 376 pairs of MZ twins.
Materials and Methods
Cohort
After quality control filtering (described below), 376 pairs of MZ twins — 169 male and 207 female — were available for CNV calling. The genotyping and initial cleaning of these samples are described at length elsewhere (Medland et al., Reference Medland, Nyholt, Painter, McEvoy, McRae, Zhu and Martin2009). Briefly, all samples were genotyped on Illumina Human610-Quad arrays. Samples with greater than 5% missing data were excluded from further analysis. MZ twin status and sex was confirmed using the SNP genotypes.
CNV Calling
The primary CNV calls were made using the software QuantiSNP v2 (Colella et al., Reference Colella, Yau, Taylor, Mirza, Butler, Clouston and Ragoussis2007) using the default settings (L = 2 M, 10 EM iterations) and using the GC correction option. A second set of CNV calls was made using PennCNV (Wang et al., Reference Wang, Li, Hadley, Liu, Glessner, Grant, Hakonarson and Bucan2007), also using the GC correction model (Diskin et al., Reference Diskin, Li, Hou, Yang, Glessner, Hakonarson and Wang2008).
Poor quality samples were excluded from the study if they failed any of the stringent quality controls metric from either CNV calling program. For QuantiSNP, samples with more than 200 CNV calls (without filtering on log Bayes factor scores), an outlier rate > 0.0075, spread of log R ratio values > 0.2 or measured spread of distribution of B allele frequencies for heterozygote genotypes > 0.07 were excluded. Further samples were excluded based on PennCNV output with greater than 70 CNV calls, a log R ratio standard deviation > 0.28, the B allele frequency standard deviation > 0.045, and an absolute waviness factor > 0.02. Thresholds for each measurement were chosen by examining their distribution for the obvious outliers. Overall, ~6% of samples failed one of these stringent quality metrics, resulting in 46 out of the original 422 twin pairs being excluded from the final dataset (due to at least one of the pair failing quality control). A strong overlap was observed between the samples excluded using each metric for both CNV calling programs.
CNV Differences within MZ Pairs
Differences within MZ pairs were first detected using QuantiSNP CNV calls, followed by confirmation of the CNV calls using PennCNV. QuantiSNP CNV calls were filtered to have at least five probes, a length of 1 kb and to be autosomal. As CNVs in highly repetitive regions — in particular centromeric regions — are known to have high false positive rates, any CNV that was annotated as having greater than 70% of the region as repetitive DNA in the UCSC version hg18 RepeatMasker annotation was removed.
For each individual, all CNVs with a QuantiSNP log Bayes factor greater than 30 were investigated for presence in the QuantiSNP CNV calls in the co-twin, regardless of the log Bayes factor. CNV calls for the twin pair are considered concordant if there is any overlap of CNV calls of the same category (deletion or duplication), irrespective of the actual called copy number and the CNV end points. Discordant CNV calls were further investigated using PennCNV, confirming both the CNV call in the original individual and the lack of CNV in the co-twin. This was repeated using a log Bayes factor threshold of 10 for the initial QuantiSNP CNV calls.
Results
CNVs in the MZ twins were called using QuantiSNP v2 on data from Illumina Human610-Quad arrays. After stringent quality control filters were applied, 376 pairs of MZ twins remained. All CNVs called with a log Bayes factor of greater than 30 were investigated for presence in the co-twin, regardless of their significance, providing 126 candidate regions for copy-number differences within the MZ pairs. A large number of these differences were repeated across other pairs and also seen at high frequency in a larger cohort (data not shown), indicating they were false positive CNVs. This was confirmed by noting that many of these CNVs fell in centromeric and other highly repetitive DNA regions that are known to generate false positive CNV calls (Wang et al., Reference Wang, Li, Hadley, Liu, Glessner, Grant, Hakonarson and Bucan2007). Based on CNVs in centromeric regions, any CNV across a region with greater than 70% of the sequence in repeat regions was removed, reducing the number of candidate regions to 16.
The remaining 16 regions were further investigated using CNV calls from PennCNV (Wang et al., Reference Wang, Li, Hadley, Liu, Glessner, Grant, Hakonarson and Bucan2007) and all called CNV regions were confirmed. For 9 of the 16 CNVs, PennCNV also identified a corresponding CNV in the co-twin. All the CNVs identified in the co-twin had a low confidence score compared to that in the primary twin, and many were called to have a shorter length or as part of a number of CNVs across a region, indicating likely false positive differences due to limitations of the calling algorithms. Additionally, one CNV identified by QuantiSNP was called as part of a larger CNV by PennCNV. In the co-twin, a CNV was found that partially overlapped the larger CNV but did not overlap the original CNV call, and so this was excluded from further analysis.
The details of the remaining six identified CNV differences are given in Table 1. There was strong evidence for one duplication on chromosome 5 and the measured log R ratios and B allele frequencies are given in Figure 1. This CNV covered 37 probes on the genotyping array and was highly significant, with a QuantiSNP log Bayes factor of 54.1 and PennCNV confidence score of 77.6. The log R ratio in the region of the insertion is slightly raised above zero, although not to the extent of the expected value of log2(3/2) (= 0.58) if three copies were present in all cells. The B allele frequencies for the heterozygous SNP clearly show two bands that indicate a duplication, but deviate towards 0.5 from their expected values of 0.33 and 0.67, indicating that this duplication is potentially a mosaic. Similar plots of log R ratio and B allele frequencies for the five identified deletions are given in supplementary Figures S1–S5. All of these deletions show the requisite reduction in log R ratio values, but none to the extent of the expected value of -1. The longer deletion on chromosome 2 (supplementary Figure S1) covers an immunoglobulin region, which is known to generate false positive CNV calls (Wang et al., Reference Wang, Li, Hadley, Liu, Glessner, Grant, Hakonarson and Bucan2007). The region of chromosome 17 identified as having a putative deletion (supplementary Figure S2) was in a region of (monomorphic) CNV probes only, so B allele frequency cannot be used as a confirmatory measure and we rely on intensity data alone. Also, the log R ratios in the surrounding regions are relatively noisy, indicating a difficult region to call CNVs using these SNP arrays. The called deletion on chromosome 15 (supplementary Figure S3) is not well supported by the B allele frequencies in the region. While these do not cluster around 0.5, the B allele frequency values are consistent across the co-twins indicating the copy-number state does not differ within the pair. The two remaining CNV (supplementary Figures S4 and S5) are both less than 10 KB and do not have any heterozygous SNPs in the region to confirm that the deletion is present only in one twin using differences in B allele frequencies. The validity of these CNV differences would require further investigation using alternative technology. Overall, after stringent quality control and manual inspection of CNV differences within MZ twin pairs, we only find strong evidence for an ~130 KB duplication on chromosome 5.
TABLE 1 Details of the Six CNV Differences Identified Across Six Different MZ Pairs by Both QuantiSNP and PennCNV

*start – stop, # probes, log Bayes factor/confidence score. The copy-number variant showing the strongest evidence for differences within an MZ pair is highlighted in bold type.
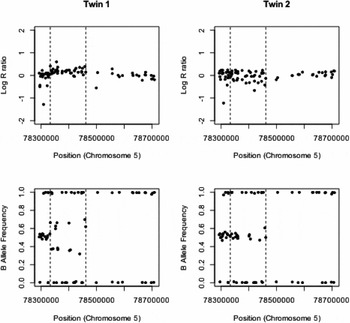
FIGURE 1 Copy-number discordance within a MZ twin pair. A ~130 KB duplication is observed between 78,333,027–8,460,944 bp (indicated by dashed lines) on chromosome 5 in twin 1. This duplication shows the characteristic increase in log R ratio above the value of zero expected when two copies of the chromosome are present. There is also clear evidence of the characteristic four genotypes (AAA, AAB, ABB, and BBB) that are seen in a duplication event in the plot of the B allele frequencies (bottom). No evidence of a duplication is observed in the co-twin (right).
As the initial threshold of a QuantiSNP log Bayes factor of greater than 30 may result in false negative CNV calls, the CNV filtering procedure was repeated with a threshold of 10. This identified 1,300 potential CNV differences within the MZ pairs, with 586 remaining after filtering out regions in repetitive sequence. Of these 586 CNVs, 499 were also called in the same individual in PennCNV. PennCNV also called an overlapping CNV in the co-twin in 107 of these CNV calls, leaving a total of 392 CNV differences called by both methods, consisting of 307 deletions and 85 duplications. Manual inspection of the 64 CNV having a log Bayes factor >20 showed similar patterns to the CNVs in supplementary Figures S1–S5, where the log R ratio in CNV region was not strongly deviated from zero and the same region in the co-twin showed similar patterns for both log R ratio and B allele frequency measurements indicating that there was no genuine difference in copy-numbers identified within the twin pairs.
Discussion
We have performed a survey of copy-number differences within 376 pairs of MZ twins using Illumina SNP arrays. Overall, there was evidence for only one copy-number difference within a pair of MZ twins, consisting of a ~130 KB duplication. Other regions had CNVs called by both QuantiSNP and PennCNV in a single member of the twin pair, but close inspection of the log R ratio and B allele frequency measurements in the co-twin indicated likely false negative CNV calls generating spurious discordance.
The use of SNP arrays for CNV calling is a relatively noisy process, with even the best calling algorithms suffering from moderate false positive and negative rates (Dellinger et al., Reference Dellinger, Saw, Goh, Seielstad, Young and Li2010; Pinto et al., Reference Pinto, Darvishi, Shi, Rajan, Rigler, Fitzgerald and Feuk2011). In this study, the concordance between two CNV calling software packages, QuantiSNP v2 and PennCNV, was checked for all CNV calls. After filtering out CNVs called in highly repetitive regions, the two CNV calling algorithms demonstrated a strong concordance, particularly for CNVs called with high confidence, indicating a low false positive rate. However, the false negative rate appears high, with a number of apparent CNV differences between MZ co-twins found by QuantiSNP v2 being rejected on the basis of the CNV also being called in the co-twin with PennCNV. Furthermore, manual inspection of intensity levels in the regions of putative CNV discordance indicated that many CNVs were not called by either piece of software. Inspection of the concordant CNV calls within MZ twin pairs shows, as would be expected, that longer CNVs are called more accurately. Thus, despite the limitations of CNV calling from SNP arrays, it is possible to conclude that large copy-number differences within unselected MZ twin pairs are rare. Different technology will be needed to accurately assess small CNVs differing within an MZ pair.
The one CNV with strong evidence for a discordance in an MZ pair was a ~130 kb duplication between 78,333,027–78,460,944 bp on chromosome 5. This duplication demonstrated the expected four bands in the B allele frequency (corresponding to genotypes AAA, AAB, ABB, and BBB), although the two heterozygous classes deviated towards 0.5 from their expected values of 0.33 and 0.67. The log R ratio also is closer to zero than would be expected for a duplication. This indicates that the duplication is probably a mosaic, in that not all cells in the individual contain it. Given the CNV is only seen in one of the twin pair, it must have occurred post fertilization. While the copy-number data are most parsimoniously consistent with mutation post embryo splitting, we are unable to rule out mutation pre-splitting with the ‘non-carrier’ co-twin either being a genuine non-carrier or being a mosaic below detection levels. Regardless of whether the CNV occurred pre- or post-splitting, it would be highly likely that cells with and without the duplication are present in the carrier, as is consistent with the observed log R ratio and B allele frequencies. As low rates of mosaicism have been observed in the general population (Jacobs et al., Reference Jacobs, Yeager, Zhou, Wacholder, Wang, Rodriguez-Santiago and Chanock2012; Laurie et al., Reference Laurie, Laurie, Rice, Doheny, Zelnick, McHugh and Weir2012), it is not possible to attribute the splitting of the embryo to this duplication.
The lack of evidence for frequent CNV differences within unselected MZ pairs supports the continued use of discordant MZ pairs to investigate the developmental etiology of disease. Several studies have provided evidence for copy-number differences within pairs of MZ twins discordant for disease (Bruder et al., Reference Bruder, Piotrowski, Gijsbers, Andersson, Menzel, Sandgren and Dumanski2008; Ehli et al., Reference Ehli, Abdellaoui, Hu, Hottenga, Kattenberg, van Beijsterveldt and Davies2012), the implication being that these are causal variants. This study demonstrates that such differences are rare in a population-based sample of MZ twins, raising the chance that the identified differences are causal. However, given that such CNV differences do occur in a population sample and genuine replication of a CNV difference in an independent pair is likely to be rare, it is important to remain cautious in conclusions about causality and to pursue other biological support for the role of a CNV region in disease. Epigenome association studies that look for discordant DNA methylation in pairs of MZ twins that are discordant for disease also implicitly make an assumption of no genetic differences within the twin pair. While this study shows that such an assumption will hold for a general population sample, the frequency of copy-number differences in pairs selected for disease discordance remains to be determined and whether frequency may vary between diseases.
Acknowledgments
We thank our twin sample for their participation; Marlene Grace and Ann Eldridge for sample collection; Anjali Henders, Megan Campbell, Lisa Bowdler, Steven Crooks, and staff of the QIMR Molecular Epidemiology Laboratory for DNA sample processing and preparation; Kerrie McAloney for study co-ordination; and Harry Beeby, Daniel Park, and David Smyth for IT support. This work was supported by grants from the Australian Research Council (ARC: A7960034, A79906588, A79801419, DP0212016, DP0343921, DP0664638, DP1093900, FT0991360) and the Australian National Health and Medical Research Council (NHMRC: Medical Bioinformatics Genomics Proteomics Program, 389891). P.M.V. and G.W.M. are supported by the NHMRC Fellowship Scheme.
Supplementary Material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/thg.2014.85.




