TwinsUK is the largest adult twin registry in the UK and is one of the most deeply phenotyped and genotyped datasets in the world. It provides a multidisciplinary platform to research both health- and social-related questions, with the overarching aim of understanding the etiology of complex disease and the aging process. The registry was started in 1992, with the initial intention to investigate osteoporosis and osteoarthritis. Such conditions are highly prevalent in women, and consequently, several hundred middle-aged women were recruited and formed the core of the initial register. Success from these early studies led to a rapid expansion of TwinsUK, and to date the cohort consists of 14,000 community-dwelling twins, male and female, aged over 18 years. Current research areas of interest include the genetics of metabolic syndrome, cardiovascular disease, the musculoskeletal system, sensory impairment and aging, as well as how the microbiome affects human health. Details of the registry’s progression have been described previously (Moayyeri et al., Reference Moayyeri, Hammond, Hart and Spector2013; Spector & Williams, Reference Spector and Williams2006). To date, the TwinsUK registry has contributed to over 850 publications and 800 international collaborations. More detailed description of research outputs may be accessed through the study website: http://www.twinsuk.ac.uk.
The Collection
Over the last 27 years, the TwinsUK registry has been enhanced through over 80 studies, some of which have been repeated over time. This has resulted in clinically rich, longitudinal phenotype information (Table 1), which may be categorized into four distinct time points (Verdi et al., Reference Verdi, Lachance, Bowyer, Abbasian, Yarand, Paraskevi and Steves2019). Recruitment strategies have predominantly involved media campaigns. These have offered opportunities for adult twin pairs to join the registry and participate in unspecific research investigating various common diseases, without selecting for particular diseases or traits. At baseline (1992–2004), over 7000 twins responded to annual questionnaires, and approximately 5500 twins attended a full comprehensive clinical visit, which included several project-led studies. Age-matched characteristics of these volunteer twins were found not to differ from a singleton population-based cohort of British women (Chingford study; Andrew et al., Reference Andrew, Hart, Snieder, de Lange, Spector and MacGregor2001), apart from a lifelong lower weight in monozygotic (MZ) twins of approximately 1 kg. The follow-up visit occurred between April 2004 and May 2007, in which 3725 twins in the registry attended a full day clinical visit, and an additional 1299 twins posted blood taken via their GPs for DNA sampling. Participants ranged aged between 18 and 82 years (mean 52.5 ± 13 years) and the majority were female (89%). Protocols for the baseline and initial follow-up visit have been described previously (Spector & Williams, Reference Spector and Williams2006).
Table 1. TwinsUK registry update

FF, female–female; MM, male–male; FM, female–male.
The second wave of follow-up visits (August 2007–April 2012) aimed to investigate the aging process; Healthy Ageing Twin Study (HATS). Inclusion criteria were women aged ≥40 years with at least one previous clinical visit (n = 4610). In total, 3125 women (mean age 59.6 ± 9 years) attended the clinical visit. Follow-up time between first and last visits ranged between 6.1 and 17.4 years, with over 600 of the participants having 4 or more previous clinical visits. HATS outcomes have previously been described (Moayyeri et al., Reference Moayyeri, Hammond, Valdes and Spector2012), including details of data collection (Moayyeri et al., Reference Moayyeri, Hammond, Hart and Spector2013).
The third wave of follow-up visits (May 2012 – May 2018) was performed to understand the interactions in disease processes between genes and the environment, as part of the Biomedical Research Centre (BRC) study. All participants of the TwinsUK registry were invited to attend a comprehensive clinical visit, which included collection of bone density/whole-body scan, cognitive and lung function, hearing and eye tests, fitness assessment (gait speed, chair stands and grip strength) and collection of blood, urine, stool and salivary samples. In total, 6686 clinical visits were made, with 3620 volunteers attending at least once and 1531 volunteers attended the clinic on 2 occasions with an average of 4 years between visits. In addition to the clinical visit, 6300 questionnaires were returned, complementing clinical data collected during the visit. Since February 2019, a further wave of follow-up visits has commenced that aims to continue the longitudinal data collection and adds further dynamic phenotyping and blood measurement over a 6-h visit incorporating standardized meals.
Longitudinal Data
Detailed clinical and biochemical phenotypes have been collected using harmonized protocols at each visit stage. A summary of a selection of clinical phenotypes is outlined in Table 2. In addition, questionnaire data have been collected on an annual basis and during visits, some which measure incident clinical endpoints such as cardiovascular accident, type 2 diabetes, chronic obstructive pulmonary disease, which have previously been described (Verdi et al., Reference Verdi, Lachance, Bowyer, Abbasian, Yarand, Paraskevi and Steves2019). Three main comprehensive questionnaires (‘TwinsUK Baseline Health’, ‘Baseline Core’ and ‘Longitudinal Core’) were collected between 2004 and 2018 (detailed in Table 3). These were in paper format, completed at respondents’ addresses and returned to the research facility. Over 2500 participants completed all 3 main questionnaires and 2300 completed either 2 of the main questionnaires. Furthermore, the demographic of the cohort provides an excellent resource to study aging where longitudinal changes are important to consider. Table 4 provides summaries of the key cognitive and frailty phenotypes we have acquired to explore questions in this area.
Table 2. Longitudinal data available in the TwinsUK registry participants
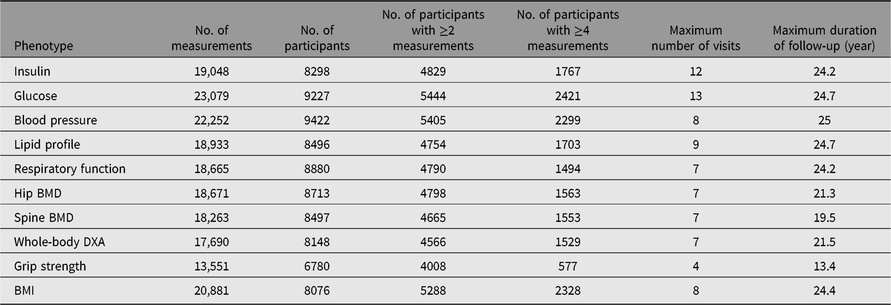
DXA, dual-energy X-ray absorptiometry; BMD, bone mineral density; BMI, body mass index.
Table 3. Longitudinal measures of main TwinsUK questionnaires

NA, not applicable.
Note: The Baseline Core and Longitudinal Core were collected 4 years apart.
Table 4. Longitudinal cognitive and frailty measures available in the TwinsUK registry participants
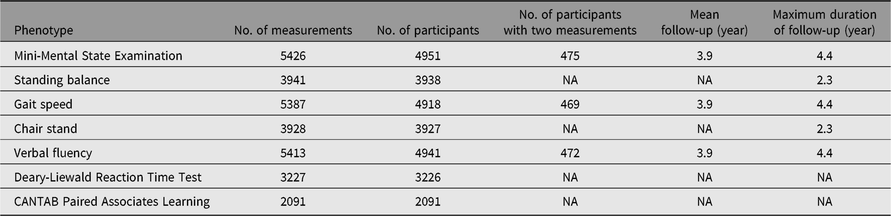
CANTAB, Cambridge Neuropsychological Test Automated Battery; NA, not applicable.
Alongside regular visits and questionnaires, TwinsUK has data linkage to official cancer and mortality data for retrospective analysis and future follow-up. Additional links to national health, education and environmental records to our own database are being established at present.
Novel Molecular and Genetic Phenotypes
In addition to epidemiological and clinical phenotypes collected from clinical visits, numerous biological samples, including body fluids (blood, urine, saliva, stool and sebum) and tissue (hair follicle, colonic mucosa, fat and skin biopsy), have been generously donated. Details of the samples collected are summarized in Table 1 and in Verdi et al. (Reference Verdi, Lachance, Bowyer, Abbasian, Yarand, Paraskevi and Steves2019). Collection methods have been described in their respective research publications, which can be found on the TwinsUK website (http://www.twinsuk.ac.uk). Here we describe the omic techniques (genomewide association studies, epigenomics, next-generation sequencing, metagenomics, metabolomics and microbiomics) that have been employed on biological samples and phenotypes from TwinsUK data. Details of some phenotypes collected prior to 2012 (e.g., telomere length) have previously been described (Moayyeri et al., Reference Moayyeri, Hammond, Hart and Spector2013).
Genomewide Association Studies
TwinsUK has contributed to many international consortia for genomewide association analysis of various phenotypes (Mills & Rahal, Reference Mills and Rahal2018). Genomewide scan data using 2 chips (Illumina HumanHap300 BeadChip and Illumina HumanHap610 QuadChip) are available for 5654 (both MZ and dizygotic [DZ]) twins. The data have been fully imputed using ‘1000 Genomes’ and ‘Haplotype Reference Consortium – (HRC)’ reference panels. TwinsUK is a member of many ongoing international consortia for meta-analysis of various traits such as height, BMI, lipids, obesity, blood pressure and back pain phenotypes. Some of the main publications from these collaborations can be found in the TwinsUK website. Our genomewide data are also being used to compile polygenic risk scores to isolate loci for various traits (Mills et al., Reference Mills, Barban and Tropf2018).
Epigenetic Markers
The first large-scale genomewide epigenetic assessment in TwinsUK was performed on DNA methylation patterns profiled on the Illumina HumanMethylation27 BeadChip in a whole blood sample of 172 female twins. This array examines 27,578 promoter CpG sites that map uniquely across the genome and some of these sites were found to be associated with age and age-related phenotypes (Bell et al., Reference Bell, Tsai, Yang, Pidsley, Nisbet, Glass and Deloukas2012). Subsequently, the Illumina Infinium HumanMethylation450 BeadChip was additionally applied up to 1000 blood samples to generate higher resolution genomewide DNA methylation profiles (Kurushima et al., Reference Kurushima, Tsai, Castillo-Fernandez, Couto Alves, El-Sayed Moustafa, Le Roy and Bell2019; Zhang et al., Reference Zhang, Spector, Deloukas, Bell and Engelhardt2015), as well as in 322 skin (Roos et al., Reference Roos, Sandling, Bell, Glass, Mangino, Spector and Bell2017) and 648 adipose (Grundberg et al., Reference Grundberg, Meduri, Sandling, Hedman, Keildson, Buil and Deloukas2013) tissue biopsy samples from twins. More recently, the Illumina Infinium MethylationEPIC array is being profiled in additional blood samples from over 400 twins. Further epigenetic datasets in TwinsUK cohort have also been generated as part of the EpiTwin study (http://www.epitwin.eu), which in collaboration with the Beijing Genomics Institute, assayed epigenomic sequencing profiles in up 5000 samples from twins aged 16–85 years. The results include methylated DNA immunoprecipitation sequencing profiles in whole blood samples from twins discordant and concordant for a wide variety of diseases and environmental exposures (Bell et al., Reference Bell, Loomis, Butcher, Gao, Zhang, Hyde and Spector2014, 2016; Davies et al., Reference Davies, Krause, Bell, Gao, Ward, Wu and Wang2014; Yuan et al., Reference Yuan, Xia, Bell, Yet, Ferreira, Ward and Spector2014).
Gene Expression Markers
During the HATS visit, 856 twins with detailed clinical profiles underwent biopsies of multiple tissues as part of the Multiple Tissue Human Expression Resource project. This was a Wellcome Trust-funded study designed to investigate gene expression across multiple tissues simultaneously with the aim of examining mechanisms involved in common trait susceptibility. Gene expression in 3 tissues and derived cells, fat, skin and lymphoblastoid cell lines (LCL) was determined using Illumina whole genome expression array (HumanHT-12 version 3) comprising 48,803 probes in 3 technical replicates (Grundberg et al., Reference Grundberg, Small, Hedman, Nica, Buil, Keildson and Spector2012). The same skin, fat and LCL RNA samples plus an additional 400 whole blood samples were RNA sequenced as part of the EuroBATS project (Biomarkers of Ageing using whole Transcriptome Sequencing) a European (EU-FP7) study (Buil et al., Reference Buil, Brown, Lappalainen, Vinuela, Davies, Zheng and Dermitzakis2015).
Whole Genome Sequencing
Whole genome sequencing (WGS) of 2000 healthy, deeply phenotyped twins formed part of the UK10K project, which used state-of-the-art next-generation sequencing methods to uncover rare genetic variants associated with health and disease. The data have been used extensively to describe population structure and functional annotation of rare and low-frequency variants (UK10K Consortium et al., Reference Walter, Min, Huang, Crooks, Memari and Zhang2015); further details can be accessed at: http://www.uk10k.org. In addition, approximately 1000 exome sequences at 30–60 × depth have been ascertained as part of projects with GoT2D consortium and Pfizer Inc. More recently, WGS of >30× coverage was carried out through collaboration with Human Longevity, Inc. (HLI) for 2377 individuals from the TwinsUK cohort. DNA samples were sequenced on an Illumina HiSeqX sequencer using a 150-base paired-end single-index read format. The data have been used to disentangle to contribution of rare variants to the blood metabolome (Long et al., Reference Long, Hicks, Yu, Biggs, Kirkness, Menni and Telenti2017), and are now under investigation to identify rare variants associated with complex diseases and traits, and for the inference of structural variants.
Metabolomics Profile
Fasting circulating metabolites levels (serum/plasma) have been assessed on TwinsUK study participants using different platforms: (1) Biocrates AbsoluteIDQ (163 metabolites for 1052 twins) (Menni, Zhai et al., Reference Menni, Zhai, MacGregor, Prehn, Römisch-Margl, Suhre and Valdes2013); (2) Metabolon Inc. (Research Triangle Park, NC, USA; 280 known metabolites and 175 unknown metabolites for 6055 twins, 591 known and 165 unknown for 2069 twins at 3 time points); (3) Nightingale Health Ltd. (27 metabolic traits, 143 metabolite concentrations, 80 lipid ratios, 3 lipoprotein particle sizes and a semiquantitative measure of albumin for 2000 twins at 3 time points). Genomewide association studies have identified several genes involved in metabolic individuality (Long et al., Reference Long, Hicks, Yu, Biggs, Kirkness, Menni and Telenti2017; Shin et al., Reference Shin, Fauman, Petersen, Krumsiek, Santos, Huang and Soranzo2014), as well as many health traits (see TwinsUK publications list: http://twinsuk.ac.uk/our-research/publications/). Nightingale Health Ltd. (Helsinki, Finland; previously known as Brainshake Ltd.) is a targeted Nuclear magnetic resonance spectroscopy platform that has been extensively applied by us and others for biomarker profiling in epidemiological studies (Barrios et al., Reference Barrios, Zierer, Würtz, Haller, Metspalu, Gieger and Menni2018). More recently, metabolomics profiling (Metabolon, Inc.) has been conducted on fecal (n = 1016; Zierer et al., Reference Zierer, Jackson, Kastenmüller, Mangino, Long, Telenti and Menni2018) and salivary samples (Nag et al., Reference Nag, Kurushima, Bowyer, Wells, Weiss, Pietzner and Steves2019).
Glycans
Glycosylation is the most common form of posttranscriptional protein modification and it is a putative mechanism in the modulation of the inflammatory response. The technology to assess glycosylation has recently become high throughput, and glycosylation of immunoglobulin G has been measured on 4900 twins while N-glycans in human serum glycoproteins have been measured in 1800 twins. Using this, we have found that glycans are highly heritable (Menni, Keser et al., Reference Menni, Keser, Mangino, Bell, Erte, Akmačić and Valdes2013) and we have been the first to observe a number of associations between glycans and important age-related traits (Barrios et al., Reference Barrios, Zierer, Gudelj, Štambuk, Ugrina, Rodríguez and Menni2015; Menni, Gudelj et al., Reference Menni, Gudelj, MacDonald-Dunlop, Mangino, Zierer, Bešić and Valdes2018).
Microbiome
Alongside the BRC study (third follow-up), over 5000 fecal samples have been collected for microbiome analysis. Twin volunteers provided stool samples, stored on site (St Thomas’ Hospital, London) at −80°C. DNA extraction and 16S rRNA sequencing using the V4 variable region of nearly 3000 samples have been completed in collaboration with Cornell University using a multiplexed approach on the Illumina MiSeq platform. Smaller subsets of twins have also been sequenced with complementary methods by the BRC Genomics Facility at King’s College London. In addition, plain saliva (700) and midstream urine (1600) specimens have undergone similar 16S amplicon sequencing using the same primers in collaboration with University of California San Diego and Stanford University.
Diversity metrics, taxonomic levels from genus through to phylum and relative abundances of operational taxonomic units (OTUs) have been used to assess microbiota associations within the TwinsUK data. Associations have been observed with a number of health deficits and medication usage (Jackson, Goodrich et al., Reference Jackson, Goodrich, Maxan, Freedberg, Abrams, Poole and Steves2016, Jackson et al., Reference Jackson, Verdi, Maxan, Shin, Zierer, Bowyer and Steves2018; Le Roy et al., Reference Le Roy, Beaumont, Jackson, Steves, Spector and Bell2017), and age-related traits, including frailty and cognition (Jackson, Jeffrey et al., Reference Jackson, Jeffery, Beaumont, Bell, Clark and Steves2016; Verdi et al., Reference Verdi, Jackson, Beaumont, Bowyer, Bell, Spector and Steves2018), among others (Menni, Lin et al., Reference Menni, Lin, Cecelja, Mangino, Matey-Hernandez, Keehn and Valdes2018). In addition, microbiota associations with diet (Menni, Jackson et al., Reference Menni, Jackson, Pallister, Steves, Spector and Valdes2017; Menni, Zierer et al., Reference Menni, Zierer, Pallister, Jackson, Long, Mohney and Valdes2017; Ni Lochlainn et al., Reference Ni Lochlainn, Bowyer and Steves2018) and socioeconomic status (Bowyer et al., Reference Bowyer, Jackson, Le Roy, Ni Lochlainn, Spector, Dowd and Steves2019) have been found. More recently, amplicon sequence variants, also known as exact sequence, have been generated from 3345 stool samples. This approach offers a higher resolution than the OTU, allowing for greater sensitivity and specificity in identifying the taxonomic associations with traits (Wells et al., Reference Wells, Williams, Matey-Hernandez, Menni and Steves2019).
Metagenomics
Whole metagenomic shotgun sequencing (WMGS) has been performed on fecal samples in 2 batches comprising 250 and 1004 volunteers from the TwinsUK registry. This larger dataset, including 161 MZ twin pairs, 201 DZ twin pairs and 280 singletons generated an average of 39M high-quality microbial reads per sample. Taxonomic and functional information have been inferred from the WMGS data. These results are being studied to determine the influence of the microbiome on the fecal and host metabolome, and to identify bacterial species and function mediating microbiome-associated increased risk for common disease.
Dietary Phenotypes
TwinsUK has detailed datasets on dietary habits, which have been collected since the inception of the registry. Data vary and include dietary indices on >5000 participants (e.g., Mediterranean Diet Score, Healthy Eating Index — 2010 and the Healthy Food Diversity Index; Bowyer et al., Reference Bowyer, Jackson, Pallister, Skinner, Spector, Welch and Steves2018). Dietary patterns, which are measured by category of foodstuff, have also been assessed through a Food Frequency Questionnaire previously used in the EPIC Study (Bingham et al., Reference Bingham, Welch, McTaggart, Mulligan, Runswick, Luben and Day2008). For details of collection, see Table 5.
Table 5. Food Frequency Questionnaire collection
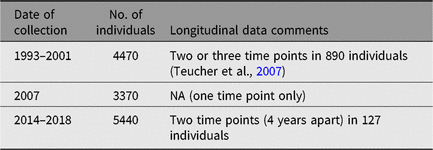
NA, not applicable.
Socioeconomic Data
The historical research focus of TwinsUK has shaped the main demographic of the twin cohort having middle socioeconomic status and education typical of a volunteer group (Moayyeri et al., Reference Moayyeri, Hammond, Hart and Spector2013; Steves et al., Reference Steves, Jackson and Spector2013). Socioeconomic status of the twin volunteers has been collected since the registry’s inception through self-reported questions (e.g., highest educational qualification status). More recently, the Index of Multiple Deprivation (IMD) has been compiled for all volunteers having UK postal codes, and data are to be linked to national databases for retrospective and future collection.
Index of Multiple Deprivation
Datasets from online government data repositories were combined, representing four of the UK’s administrative countries: England (IMD version 2015), Scotland (IMD version 2016), Wales (IMD version 2014) and Northern Ireland (IMD version 2017). The IMD is a composite measure of area-level deprivation and considers the following domains: income, employment, education, skills and training, health deprivation, crime, barriers to housing and services, and the living environment. As methods may vary between the countries, and ranks are inappropriate (given the differing numbers of administrative districts in each country), the decile score was combined as a relative measure of deprivation. Datasets were matched to postcodes or Lower Layer Super Output Area (LSOA) codes at 17,498 time points for 12,041 individuals. Mean IMD decile score (considering all time points) was 6.49.
Future Directions and Collaborations
Longitudinal and detailed clinical, biochemical, behavioral, socioeconomic and deep omics (including multitissue characterization) of participants for nearly 30 years has provided a unique resource to study complex diseases and domains of healthy aging in the TwinsUK population. These, in conjunction with novel dynamic testing at study visits and lifestyle intervention studies, offer a unique opportunity to explore personalized medicine. High-quality data collection, database management, biological sample storage and statistical quality control enhance the resource. In addition, a key strength of the resource lies in the highly engaged and loyal population; this is evident from the high retention levels of participation across studies. Blood, urine, DNA and multiple tissue samples are available for future measurements. Online questionnaires and active engagement with our twin participants using text messages, emails and social networking enable responsive and agile data collection. Our Volunteer Advisory Panel is key to developing new strategies and governance of participants, informing on decisions about the ethics, practicalities and appropriateness of potential studies.
The TwinsUK registry has a history of numerous successful scientific collaborations, and we remain committed to providing the scientific community with access to the phenotype data from the TwinsUK Resource. TwinsUK has an exemplary record for data sharing with over 800 data access requests, 150,000 samples shared to over 100 collaborators and over 600 publications in the past 6 years. Detailed descriptions for researchers of data and samples are on the data access pages of the website (http://www.twinsuk.ac.uk/data-access/cohortdata-description/); here, over 10,000 phenotypes can be searched. Longitudinal population studies funding from the Wellcome Trust continues to fund the core functions of TwinsUK and opens up the resource to successful cross-cohort collaborations. Over the next 5 years, TwinsUK will integrate electronic health records into an enhanced deep tissue omics resource and continue dynamic phenotypic testing into clinical visits. In addition, we will extend the age range of the registry to include volunteer twins from birth to adulthood, thus opening up the resource to study unique twin gene–environment interactions across the life course. New efficient broad consent will ensure that the communication with participating twins is ethical and proportionate. New annual sociological questionnaires will harmonize with English Longitudinal Study of Ageing and other LPS (1946/1958). We will also standardize mental health phenotypes between the complementary Twins Early Development Study (TEDS) such that, together, TwinsUK and TEDS cohorts will be an unparalleled twin resource across the life course. These developments will ensure TwinsUK will be a unique global resource of longitudinal omics and twin research across the life course, with immense potential for future scientific exploitation.
Acknowledgments
TwinsUK is core funded by the Wellcome Trust, and grants WT081878MA and WT202786/Z/16/Z contributed to the majority of data described. In addition, TwinsUK receives funding from Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.







