Asthma is a common disease that continues to increase in incidence. It is characterized by reversible narrowing of the airways, often associated with allergic inflammatory processes in the airway wall.
Asthma is also a prototypical complex genetic disease. A strong familial component to the occurrence of asthma has been recognized since the time of Cooke (Cooke & Vander Veer, Reference Cooke and Vander Veer1916), and genome wide association studies (GWAS) have now identified over 60 individual risk loci (Demenais et al., Reference Demenais, Margaritte-Jeannin, Barnes, Cookson, Altmüller, Ang and Nicolae2018; Pividori et al., Reference Pividori, Schoettler, Nicolae, Ober and Im2019).
One classic approach to genetic disease has been to study isolated populations where the total number of causative loci is likely to be smaller, with risk alleles at higher frequency due to inbreeding and genetic drift, and exposure to environmental risk factors more homogenous (Kenny et al., Reference Kenny, Kim, Gusev, Lowe, Salit, Smith and Pe’er2011; Sheffield et al., Reference Sheffield, Stone and Carmi1998). The island of Tristan da Cunha offers such a resource for the study of asthma: asthma prevalence is high and the present population is descended from only a small number of founding members. We have previously described (Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996) a complete population survey of asthma on the island, and here we report the strength of association of asthma with a panel of asthma and atopic disease risk variants previously identified in large outbred population studies (Moffatt et al., Reference Moffatt, Gut, Demenais, Strachan, Bouzigon and Heath2010; Paternoster et al., Reference Paternoster, Standl, Chen, Ramasamy, Bønnelykke and Duijts2011; Sleiman et al., Reference Sleiman, Flory, Imielinski, Bradfield, Annaiah, Willis-Owen and Hakonarson2010).
Methods
Asthma phenotype measurements and blood samples were obtained from the inhabitants of Tristan da Cunha, an isolated island in the South Atlantic (Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996). The entire island population at the time of examination (1991 and 1996) formed a single large extended family descended from 28 original founders. Settlement of Tristan da Cunha occurred beginning in 1817 with soldiers who remained behind when a British garrison was withdrawn from the island, followed by the survivors of several shipwrecks. In 1827, five women from St Helena, one with children, emigrated to Tristan da Cunha and married island men. William Glass and his wife of African descent are said to have been asthmatic and could be the origin of a genetic founder effect for asthma in this population. Inbreeding has resulted in kinship resemblances of at least first cousin levels for all individuals (F = .04). A high prevalence of asthma has been noted on the island over at least 80 years, as well as on St Helena, where many residents are related to the Tristan da Cunha population.
The Tristan da Cunha family pedigrees were ascertained through review of baptismal, marriage and medical records, as well as reliably accurate historical records of the early inhabitants (Roberts, Reference Roberts1968; Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996). The prevalence of asthma on Tristan da Cunha has been reported to be high in several reports (Black et al., Reference Black, Thacker, Lewis and Thould1963; Wild, Reference Wild1923; Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996) spanning 80 years — Macklin comments on ‘a condition they call “ashmere”, meaning asthma. This seems to be the most prevalent complaint on the island’ (Wild, Reference Wild1923, p. 233). In the present population, one-third met strict criteria for diagnosis of asthma.
Clinical Characterization
Standardized questionnaires based on that of the American Thoracic Society were used to record the presence of respiratory symptoms such as cough, sputum and wheezing; the presence of other chest disorders, including recent upper respiratory tract infection, allergic history; asthmatic attacks, including age at onset, offset, confirmation by a physician, prevalence, severity and precipitating factors; other illnesses and smoking history; and all medications used within the previous 3 months. A physician-confirmed asthmatic attack was the principal criterion for diagnosis of asthma.
Skin atopy was determined by skin prick tests to common allergens: Aspergillus fumigatus, Cladosporium, Alternaria, egg, milk, wheat, tree, dog, grass, horse, house dust, cat, feathers and house dust mites Dermatophagoides farinae and Dermatophagoides pteronyssinus. Saline and histamine controls were also performed (Bencard Laboratories, Mississauga, Ontario). Antihistamines were withdrawn for at least 48 h prior to testing. Wheal diameters were corrected by subtraction of the saline control wheal diameter, and a corrected wheal size of >3 mm recorded 10 min after application was considered a positive response.
Airway responsiveness was assessed by a methacholine challenge test using the tidal breathing method (Cockcroft et al., Reference Cockcroft, Killian, Mellon and Hargreave1977). Doubling doses of methacholine from .03 to 16 mg/ml were administered using a Wright nebulizer at 4-min intervals to measure the provocative concentration of methacholine, producing a 20% fall in FEV1 (PC20). In those subjects with a baseline FEV1 (forced exhalation volume in 1 s) >70% of predicted (Crapo et al., Reference Crapo, Morris and Gardner1981), a bronchodilator response to 400-mg salbutamol aerosol was used to determine airway responsiveness. Both methacholine challenges and bronchodilator responses were measured using a computerized bronchial challenge system (S&M Instrument Co. Inc., Doyleston, PA) consisting of a software package and interface board installed in a Toshiba T185C laptop computer and connected to a flow sensor (RS232FS). The power source for instruments used on Tristan da Cunha has been described (Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996).
Increased airway responsiveness was defined as a PC20 <4.0 mg/ml or a >15% improvement in FEV1 15-min postbronchodilator. Participants were asked to withhold bronchodilators at least 8 h before testing; inhaled or systemic steroids were maintained at the usual dosage.
Genotyping
Single-nucleotide polymorphism (SNP) genotyping was carried out using the Sequenom MASSarray system. A set of 43 candidate SNPs in 37 genes was selected from the published literature, especially the meta-analysis of Moffatt et al. (Reference Moffatt, Gut, Demenais, Strachan, Bouzigon and Heath2010; see Table 1). The genotyping assay failed for two SNPs. The Human Genome Diversity Project (HGDP; Li et al., Reference Li, Absher, Tang, Southwick, Casto, Ramachandran and Myers2008) sample genotypes at the same panel of SNPs provided external controls to assess the relationship of the Tristan da Cunha population with those from other regions.
Table 1. Panel of previously reported asthma-associated SNPs tested in the Tristan da Cunha sample, and results for coxme and MCMCglmm tests of allelic association to asthma

Note: Bold type indicates the most statistically significant result.
Statistical Genetic Analysis
Descriptive statistics and plots were generated using the R Statistical Computing Environment (R Core Team, 2019). Various genetic analyses were performed using the sib-pair package (Duffy, Reference Duffy1997, Reference Duffy2019). We estimated the heritability of asthma based on the recorded pedigree under the multifactorial threshold model using the R MCMCglmm package (Hadfield, Reference Hadfield2010). A mixed-model genetic association analysis of onset of asthma under the Cox proportional hazards was performed using the R coxme package (Therneau, Reference Therneau2015; Therneau and Grambsch, Reference Therneau and Grambsch2000), including the kinship matrix based on the recorded pedigree and including the genotypes at each SNP in turn along with the first five principal components (including a quadratic fixed effect for the first principal component, PC1) from a genetic principal components analysis of the panel of candidate SNPs: Given that these are known asthma candidate variants, we also calculated a polygenic risk score (PRS) based on the effect sizes reported in the GABRIEL asthma meta-analysis (Moffatt et al., Reference Moffatt, Gut, Demenais, Strachan, Bouzigon and Heath2010).
Results
As previously described (Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996), at the time of the fieldwork for this study, the population of the Island of Tristan da Cunha was 299 individuals, of whom 282 individuals (97%) provided medical histories and underwent skin prick allergen testing. A total of 102 had a medical history consistent with a diagnosis of asthma plus significant airway hyperresponsiveness to inhaled methacholine. Among the 269 individuals successfully genotyped, 96 (36%) met these criteria. The median age of onset was 10 years (interquartile range 5–21).
For genetic analysis, these 269 individuals were incorporated into a 559-member, 10-generation pedigree. The mean inbreeding coefficient for the asthma cases was .045, and that of unaffected individuals .046. While 15% of offspring were asthmatic when neither parent was affected, this rose to 30% when one parent was affected and 50% when both parents were affected. Based on the entire pedigree, the heritability of asthma under the multifactorial threshold model (using MCMCglmm) was .51 (95% highest posterior density interval [.21, .76], see Table 2).
Table 2. Asthma heritability estimates based on pedigree and SNP-derived kinship matrices, and contribution of rs2786098 to diagnosis of asthma
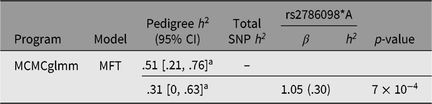
a Highest posterior density (HPD) interval.
Table 3. Results of Cox proportional hazards mixed model (R coxme package) for association between rs2786098 and age at onset of asthma
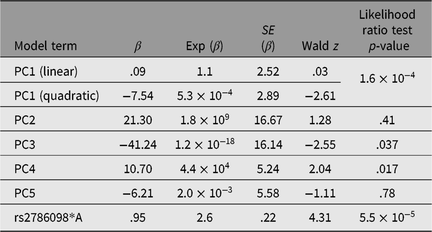
Of the 41 successfully genotyped SNPs, the five (uncommon) FLG coding variants were all monomorphic (see Table 1). The multilocus genetic distances between the Tristan and HDGP continental populations were roughly equal, with the Americas slightly closer (Europe multilocus F statistics = .16; Africa .20; Americas .13; Asia .16; see Figure 1). On plotting the relationship between asthma risk and loading on the first genetic principal component (Figure 2), a significant quadratic relationship was found, with risk greatest for those individuals intermediate between African and European mean values. This is why we chose to adjust for a quadratic effect of PC1 in the individual SNP association analyses.
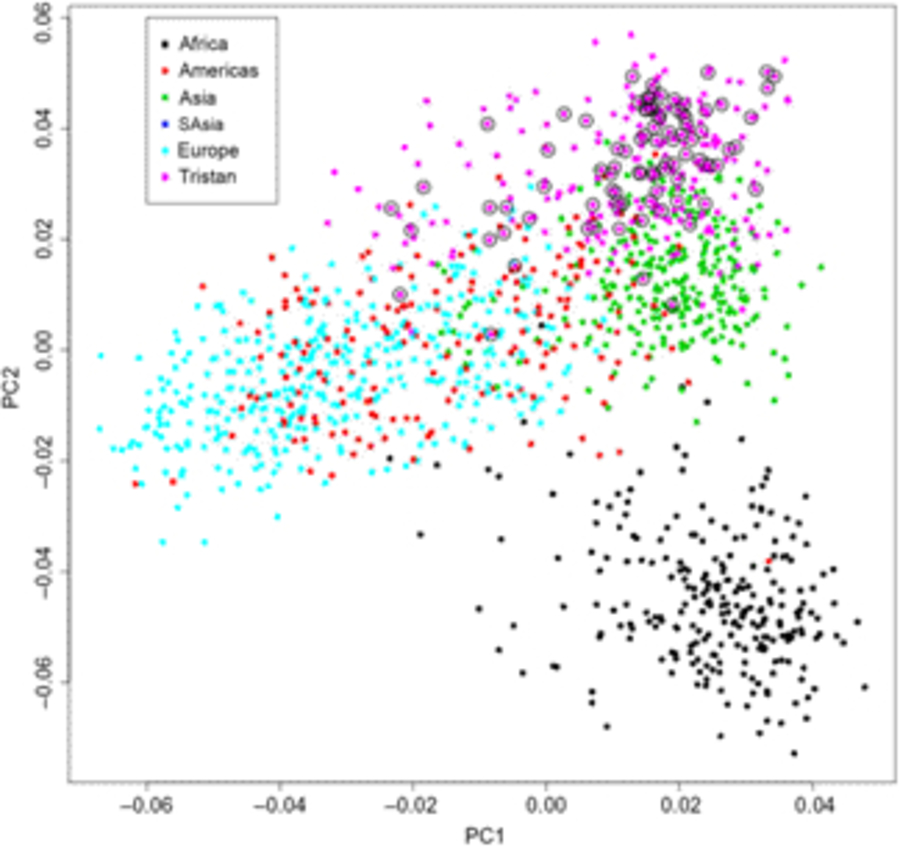
Fig. 1. First and second principal components scores from genetic principal components analysis combining Tristan da Cunha and 1000 Genomes comparison samples for the 43 SNPs. Open circles designate asthma cases.
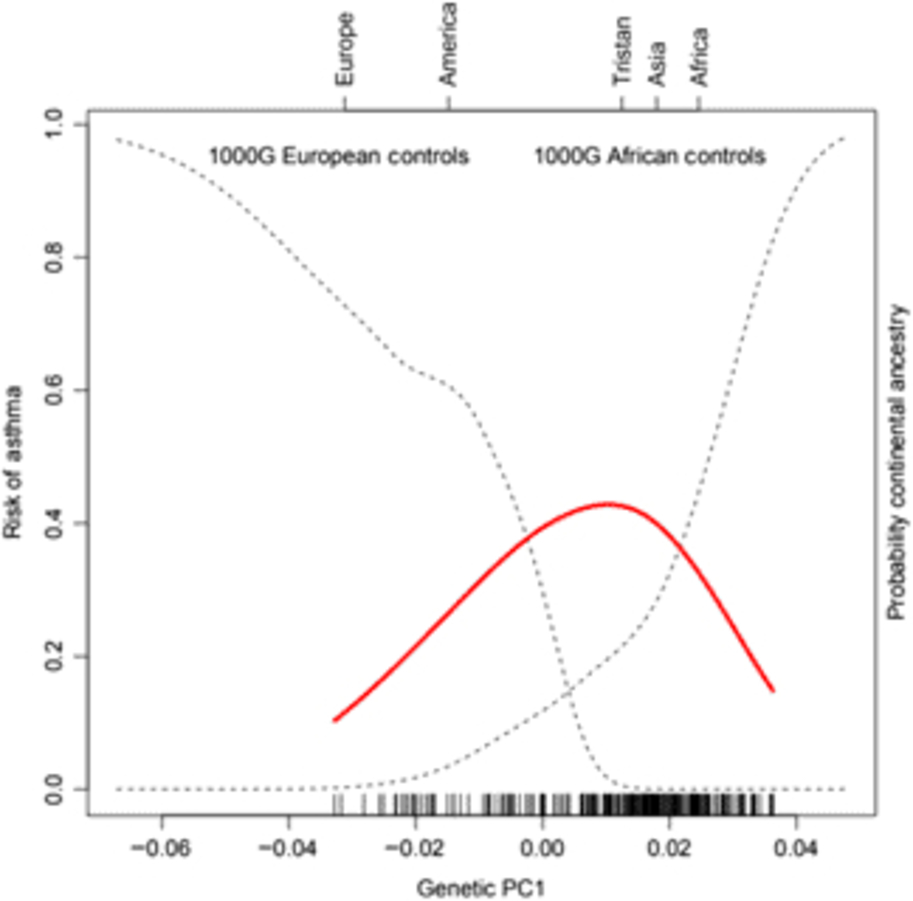
Fig. 2. (Colour online) Risk of asthma (red line) versus average ancestry as measured by first genetic principal component (PC1) for the SNP panel. The mean location of the 1000 Genomes external control populations on PC1 is indicated at the top of the plot, and the two dotted curves represent the probability of membership of the European and African 1000 Genome samples for values of PC1. The smooth non-linear fit for asthma risk is from a localized regression.
Only one SNP met the Bonferroni-corrected significance threshold for association to asthma — rs2786098 (coxme crude allelic p = 5.5 × 10−5; p = .002 corrected for 37 tests, see Table 3). The allelic odds ratio for rs2786098*A was 2.6, and of 12 A/A homozygotes, 10 were asthmatics. Age at onset was 8 years in A/A homozygotes versus 19 years in C/C homozygotes, but the Age × Genotype interaction term in the mixed Cox model was not statistically significant (main effect of age, p = .06; interaction p = .60). The frequency of the rs2786098*A allele in cases was 31%, and 13% in unaffecteds (Table 4). In Western Europe, the A frequency is usually ~22%, but is ~8% in African-descended populations.
Table 4. Allele and genotype frequencies for rs2786098 near DENNDB1 on Tristan da Cunha and in comparison populations from the 1000 Genomes project
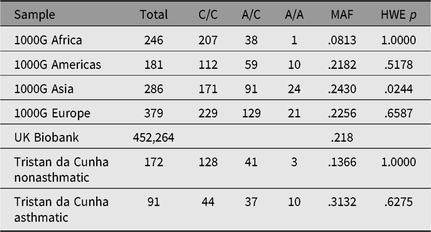
Note: MAF = minor allele frequency; HWE = Hardy–Weinberg equilibrium.
For a PRS based on all these asthma candidate genes and the estimates from the GABRIEL asthma meta-analysis, unaffected individuals had a significantly higher mean PRS than asthmatics (coxme p = .02). This is not due to rs2786098, because in the GABRIEL meta-analysis, over all studies this SNP was not associated with asthma (OR = 1.04).
Conclusions
Among the candidate SNPs for asthma that we tested, the most strongly disease-associated allele was rs2786098*A, with a large effect size (allelic OR = 2.6) in the Tristan da Cunha population. The SNP rs2786098 on chromosome 1q31 was originally reported to be associated with childhood asthma in 2010 by Sleiman et al. (Reference Sleiman, Flory, Imielinski, Bradfield, Annaiah, Willis-Owen and Hakonarson2010). The major C allele was increased in asthmatics in the European origin discovery (with an allele frequency of 8D1C 5% vs. 78% in controls) and replication (82% vs. 77%) sets, but the direction of association was reversed in African–Americans (90% vs. 93%, p = 4 × 10−5). Those authors reported the association peak included multiple additional SNP spanning an interval of 400 kbp, and flagged the DENNDB1 in this interval as a plausible candidate, given that it was known to be a TNF-α binding protein, and expressed in particular dendritic cell subtypes.
In several subsequent large GWAS meta-analyses (Demenais et al., Reference Demenais, Margaritte-Jeannin, Barnes, Cookson, Altmüller, Ang and Nicolae2018; Moffatt et al., Reference Moffatt, Gut, Demenais, Strachan, Bouzigon and Heath2010; Zhu et al., Reference Zhu, Lee, Chaffin, Chung, Loh, Lu and Liang2018), there was no evidence of association of rs2786098 to asthma. For example, analyzing 15,500 (doctor-diagnosed) asthmatics and 106,000 controls from the UK Biobank, the allelic odds ratio for rs2786098*A is .9999, with a 95% confidence interval (95% CI [.9962, 1.0036]; Canela-Xandri et al., Reference Canela-Xandri, Rawlik and Tenesa2018). Very recently, Ortega et al. (Reference Ortega, Li, O’Neal, Hawkins, Manichaikul and Barjaktarevic2019) have reported a relationship between a coding change (rs35173535) in DENND1C and acute exacerbations of chronic obstructive pulmonary disease (p = 7.4 × 10−7), which they thought to further implicate this family of genes in lung disease.
Despite these unpromising epidemiological replication results, Yang et al. (Reference Yang, Hojer, Zhou, Wu, Wuster, Lee and Chan2016) have shown that Th2 cells from rs2786098*A carriers of European ancestry produce significantly more DENNDB1 mRNA and protein than those from C/C carriers, as well as lower amounts of IL-4 and IL-13 production following T-cell receptor stimulation. These effects were replicated in rs2786098*A transfection in C/C Th2 cells and siRNA DENNDB1 knockdown in A/A Th2 cells. In Dennd1b knockout mice (close in biochemical effect to the human C/C genotype), there is a parallel increase in airway inflammation following allergen challenge. These authors thought this implied that the variable population level association between rs2786098 and asthma might reflect strong modification by covariates such as age or asthma subtype. One would expect the environment of Tristan da Cunha differs greatly in terms of risk factor exposures from that of other populations studied, even though, for example, a positive skin prick test to D. pteronyssinus house dust mite allergen was seen in 80% of the asthmatics in the sample (Zamel et al., Reference Zamel, McClean, Sandell, Siminovitch and Slutsky1996). The latter sensitization is characteristic of atopic asthmatics in most Western environments.
In the Tristan population, we do not see any age interaction effects, and the association is with the A allele that is putatively protective in European populations. Our result does not reach the usual genome wide significance threshold of 5 × 10−8, and the inclusion of this variant in our analysis was solely based on the original report of association. While the frequency of the rs2786098*A allele in our unaffected controls is close to that in the HGDP African-descended individuals, that in the cases is greater than that in HDGP European samples. We have attempted to exclude ethnic confounding using genetic principal components based on the SNP panel in our association analyses. Given the depth of the pedigree, much confounding from ethnic admixture should have broken down.
We cannot therefore definitely implicate DENND1B genotypes in the high rates of asthma in this population, but retain a high index of suspicion of a relationship given the various lines of evidence presented.
Acknowledgments
KAS is supported by a Canada Research Chair, the Sherman Family Chair in Genomic Medicine and CIHR Foundation Grant 353710.








