Introduction
Inner retinal degeneration, in particular of the retinal ganglion cells (RGCs), is a hallmark of glaucoma (Qu et al., Reference Qu, Wang and Grosskreutz2010) and of interest in many retinal diseases (Barber et al., Reference Barber, Lieth, Khin, Antonetti, Buchanan and Gardner1998; Votruba et al., Reference Votruba, Moore and Bhattacharya1998; Williams et al., Reference Williams, Morgan and Votruba2010). The study of RGC morphological and functional changes is vital in understanding disease progression and visual outcomes. While the primate provides the closest match to the human eye, the rat and mouse offer more tractable models for studying retinal degeneration.
Electrophysiological and morphological studies of single RGCs have revealed the functional and structural basis for the classification of RGCs. The matching of Boycott and Wassle's (Reference Boycott and Wassle1974) morphologically described cat α and β cells to Enroth-Cugell and Robson's (Reference Enroth-Cugell and Robson1966) functionally described cat Y and X cells highlighted the close relationship between structure and function in RGCs. The intrinsic electrical properties of the cat RGC are robust indicators of RGC type (O'Brien et al., Reference O'Brien, Isayama, Richardson and Berson2002). RGC receptive field sensitivity maps to the dendritic field as a function of bipolar cell connectivity (Brown et al., Reference Brown, He and Masland2000).
The shape and branching of the dendritic tree have been used to define the subclassification of RGCs in healthy adult and developing retina. The morphological classification of RGC types in the cat (Hochstein & Shapley, Reference Hochstein and Shapley1976; Citron et al., Reference Citron, Emerson and Levick1988; Coelho et al., Reference Coelho, Di Gesu, Lo Bosco, Tanaka and Valenti2002), primate (Peichl, Reference Peichl1991; Dacey, Reference Dacey1993), mouse (Sun et al., Reference Sun, Li and He2002b; Coombs et al., Reference Coombs, van der List, Wang and Chalupa2006), and rat (Huxlin & Goodchild, Reference Huxlin and Goodchild1997; Sun et al., Reference Sun, Li and He2002a) is comprehensive, with many cells matching functionally described types.
By contrast, RGC classification in retinal degenerative diseases where the dendritic architecture is altered remains problematic. Recent studies have reported reductions in dendritic field size and branching density in diseased RGCs (Weber et al., Reference Weber, Kaufman and Hubbard1998; Shou et al., Reference Shou, Liu, Wang, Zhou and Zhao2003; Leung et al., Reference Leung, Weinreb, Li, Liu, Lindsey, Choi, Liu, Cheung, Ye, Qiu, Chen, Yung, Crowston, Pu, So, Pang and Lam2011; Kalesnykas et al., Reference Kalesnykas, Oglesby, Zack, Cone, Steinhart, Tian, Pease and Quigley2012; Feng et al., Reference Feng, Zhao, Yoshida, Chen, Yang, Kim, Cang, Troy and Liu2013). The classification of RGCs by these same measurements (dendritic field diameter, branching density, and stratification) may be unreliable in disease states, particularly in rodent eyes where the features defining RGC classes can be subtle. Without classification, bias in the labeling of any given RGC type could show artifactual shrinkage or recovery of the dendritic tree following treatment.
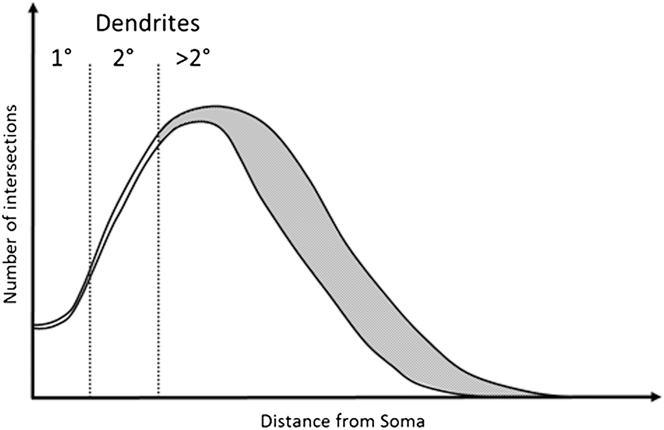
In this study we explore the possibility that a robust RGC classification system can be based on dendrite features that are relatively resistant to the effects of early disease. We base this on the observation that early to moderate RGC damage consistently affects tertiary dendrites, leaving more proximal dendritic structures intact (Weber et al., Reference Weber, Kaufman and Hubbard1998; Morgan et al., Reference Morgan, Datta, Erichsen, Albon, Boulton, Hollyfield, Anderson and LaVail2006; Williams et al., Reference Williams, Morgan and Votruba2010). This concept is illustrated in Fig. 1 based on modeling of typical Sholl profiles in normal and diseased tissue (Weber et al., Reference Weber, Kaufman and Hubbard1998; Morgan et al., Reference Morgan, Datta, Erichsen, Albon, Boulton, Hollyfield, Anderson and LaVail2006; Williams et al., Reference Williams, Morgan and Votruba2010).

Fig. 1. Example of a degenerated Sholl plot. Graphical representation of a Sholl plot from healthy and degenerate RGCs as observed in Williams et al. (Reference Williams, Morgan and Votruba2010), Weber et al. (Reference Weber, Kaufman and Hubbard1998), and Morgan et al. (Reference Morgan, Datta, Erichsen, Albon, Boulton, Hollyfield, Anderson and LaVail2006). Under moderate degeneration, Sholl plots show high deviation (gray shaded area) from healthy plots distal to the soma, with little deviation proximal to the soma. Primary and secondary dendrites occur within this proximal region and are therefore more likely preserved. Measurements of primary and secondary dendrites may therefore provide robust and stable classification criteria.
Our hypothesis is that a new set of parameters based on primary and secondary dendrite features can be used to indicate the class composition of a population of labeled RGCs from diseased retinas and act as a check against the selective labeling of cells from any given class. We therefore determined the extent to which structural features of primary and secondary dendrites could be used to separate cell types and derive a new classification system.
Materials and methods
Experiment procedures were conducted in accordance with the guidelines of the Society for Neuroscience and Home Office (UK) regulations governing the use of animals in research.
DiOlistic labeling
Twenty retired breeder Brown Norway rats, aged 4–6 months, were killed by overdose of CO2. Death was confirmed by cervical dislocation and the eyes immediately enucleated. The retinas were dissected in Hank's balanced salt solution (Life Technologies) and orientated using a cut in the nasal quadrant. Dissected retinas were flat mounted on cell culture inserts (Millipore 0.4 µm pore, Fisher Scientific) and DiOlistically labeled following a modified protocol of Gan (Gan et al., Reference Gan, Grutzendler, Wong, Wong and Lichtman2000). A Helios gene gun (BioRad) was used to fire 1.7 µm tungsten particles coated with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) and 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO) (both Life Technologies) into the retina at 689.5 kPa (100 psi) and a gun nozzle to retinal surface distance of 4 cm. Aggregated particles were excluded by placing a cell culture insert (BD Falcon 3.0 µm, BD Biosciences) between the retina and gene gun. Retinas were then cultured in Neurobasal-A medium for 30 min at 37°C. Retinas were flat mounted ganglion layer up on Histobond® coated slides (Fisher Scientific), fixed with 4% paraformaldehyde (pH 7.4) for 20 min, and washed with phosphate buffered saline (PBS). TO-PRO-3 iodide (Life Technologies) diluted 1:1000 in PBS was applied for visualization of ganglion cell and inner nuclear layers in order to measure stratification of RGCs in the inner plexiform layer (IPL). Retinas were then mounted in ProLong® gold antifade reagent (Life Technologies). A Zeiss LSM 510 confocal microscope (Carl Zeiss AG, Germany) with a x20 air objective (NA 0.8) was used to capture multichannel z-stack images (1024 × 1024 pixels, 0.54 µm/pixel, slice thickness 1 µm) of single DiI and DiO labeled RCCs, with TO-PRO-3 stained ganglion cell to inner nuclear layers. RGC distance from the optic nerve in the x and y plane was recorded using a custom scanning stage attached to the microscope.
Analysis of new RGC parameters
Only those RGCs with an axon traceable toward the optic nerve and a dendritic tree easily distinguishable from those of other cells were included. In addition, RGCs whose primary dendrite origin at the soma was obscured was not included. In the analysis, the following dendrite parameters were determined:
• The number of primary dendrites (PD)—i.e., those originating directly from the soma.
• The distance from the soma to the primary branch point was measured for each primary dendrite using the simple neurite tracer plugin (Longair et al., Reference Longair, Baker and Armstrong2011) in Fiji (Schindelin et al., Reference Schindelin, Arganda-Carreras, Frise, Kaying, Longair and Pietzsch2012) and a mean primary dendrite length (PDL) given for each cell.
• Primary branch point field (PBPF) area: In Z-collapsed images, the primary branch points were connected and the area of the resultant field measured to give the PBPF area.
• PBPF center of gravity offset (CGO) from the soma: As a measure of asymmetry in the dendritic tree, the PBPF's center of gravity offset from the soma was measured. The PBPF centroid was found and its distance from the soma center measured. This was expressed as a percentage of the diameter of the PBPF to account for RGC size variation.
• Secondary branch point field (SBPF) area.
• SBPF center of gravity offset (CGO) from the soma.
• PD asymmetry: As a further measure of asymmetry, the distribution of PD about 360° of the cell soma was measured using the oval profile plugin in Image J (NIH, US). In Z-collapsed images, a circle with a radius of 25 µm from the soma center was drawn, and the angle at which each primary dendrite intersected the circle measured. The angle formed between the primary dendrite immediately clockwise and counter clockwise of the axon was taken so that a larger angle would represent greater asymmetry in the PBPF.
• Proximal branching density: As a measure of branching density in the proximal dendritic tree, the PBPF area was expressed as a percentage of the SBPF area.
• Soma diameter: RGC soma diameter was also measured by drawing a polygon around the soma in Z-collapsed images, the resulting area was converted to a circle and the diameter calculated.
• Distance of RGC soma in the X and Y plane from the optic nerve head (ONH) center, also expressed as a vector length.
• PD cross-sectional area: As a measure of primary dendrite thickness at the origin from the soma, primary dendrite traces derived from the simple neurite tracer plugin were resliced to give a cross-sectional view and the area at the origin measured.
• PD Feret diameter: As a further measure of dendrite thickness, the minimum and maximum Feret diameters of the resliced dendrite were measured.
Comparison to existing classification
RGCs were classified according to Sun (Sun et al., Reference Sun, Li and He2002a) on the basis of dendritic field diameter, dendrite stratification within the IPL, soma diameter, and branching density. Sholl analysis using FastSholl plug-in (Gutierrez & Davies, Reference Gutierrez and Davies2007) for MATLAB (MathWorks) was conducted on all cells. Cells were grouped according to Sun (Sun et al., Reference Sun, Li and He2002a) and the mean and SD of the new parameters were calculated for each resulting group. A principal component analysis (PCA) was then performed in SPSS (version 18.0, IBM) to reveal factors driving separation and a discriminant analysis (SPSS) performed in order to reveal if the new parameters could classify into the same groupings as Sun. RGCs were randomly assigned to either a training data sample or hold-out sample. Each RGC was assigned a random number from a chi-square distribution with 1 degree of freedom using the random number generator function in SPSS. RGCs were then sorted by size of random number in ascending order with the first 30 RGCs assigned to a hold-out sample and the remaining 101 RGCs to the training sample.
Results
RGC imaging and Sun classification
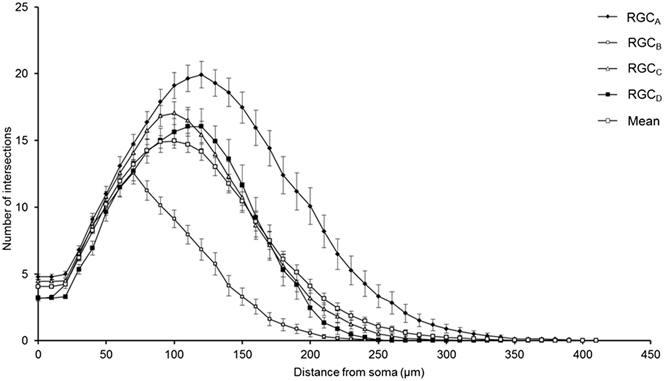
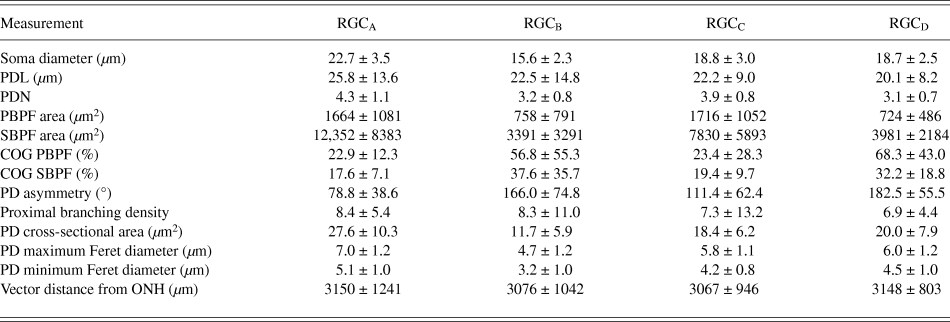
One-hundred and thirty one RGCs from 20 retinas imaged were included in the analysis based on the described exclusion criteria. The retinal locations of 114 RGCs are shown in Fig. 2 (the locations of 26 cells were not measured). The mean Sholl profile for each RGC type is plotted in Fig. 3. Classification according to the criteria of Sun et al. (Reference Sun, Li and He2002a) gave comparable percentages of cell types in the overall population, reported here as present study percentage (Sun percentage), at 24% (18%) RGCA, 29% (30%) RGCB, 32% (36%) RGCC, and 15% (18%) RGCD. Typical cells for each of the 4 classes are shown in Fig. 4. Our sample therefore provides a similar breakdown of the RGC population for the derivation of classification parameters. The mean and standard deviation according to Sun type for all classification parameters measured are detailed in Table 1.

Fig. 3. Variation of Sholl plot between RGC types. Mean Sholl plot (n = 131) and Sholl plots of RGCs grouped according to type reveal the variation within the pooled population. The plots highlight how analysis of an unclassified population of RGCs could suffer from intrinsic bias through disproportionate numbers of cell types; the leftward shift observed in degeneration is also seen when comparing healthy types. Classification is therefore critical in removing type specific biases. Error bars show SEM.

Fig. 4. RGC types. RGC types display a characteristic morphology, represented by z-compressed confocal images and tracings of typical cells of each type (Bistratified RGCD is depicted as inner and outer IPL stratifications, left and right tracings, respectively). Arrows denote axon. Scale bar = 100 µm.

Fig. 6. Discriminant analysis of RGCs. The discriminant scores of each RGC (n = 131) are plotted in X, Y dimensions corresponding to the first two discriminant functions. Here variance between groups is maximized so that group centroids (black pentagons) are at maximal distance from each other. RGC classification into a given group is achieved when distance to group centroid is shortest for that given group. The area in which this is true defines the group boundary. When labeled according to type, it can be seen that the majority of RGCs are correctly classified to their group by the discriminant functions.
Retinal eccentricity does not affect classification
One-way ANOVA with post-hoc Bonferroni showed no significant difference between RGC types with respect to retinal eccentricity when expressed as vector length (P > 0.05). This confirmed that labeling was not regionally biased. RGC distance from the ONH center expressed as distance in x, y and vector distance all showed no correlation to any variables (P > 0.05) demonstrating that retinal eccentricity had no effect on variable measurements. The classification of RGCs into types according to Sun was also unaffected by retinal eccentricity as dendritic field diameter and soma diameter showed no correlation to vector length (P > 0.05, Pearson's correlation).

Fig. 2. Distribution of recorded RGCs. Retinal eccentricity of 114 RGCs shown as distance from the ONH. RGC types show an even spread through retinal quadrants and central/peripheral retina demonstrating minimal regional labeling biases in DiOlistic delivery. Axes show distance from ONH.
RGCs differ in size and asymmetry of the proximal dendritic tree
A PCA was performed in order to determine the variables that contribute to the variation in morphology between RGC types and are thus the most useful in discriminating RGC types. PCA loses power and relevance if variables included are not correlated. RGC distance from the ONH in the x and y plane and as vector length were all discarded as variables due to the lack of correlation (as discussed above). Pearson's correlation and Spearman's rank tests were performed on normally and non-normally distributed variables, respectively. PDL and proximal branching density showed a significant correlation with fewer than half of the total variables (P > 0.05) and so were excluded from PCA. The remaining variables all showed a significant correlation (P < 0.05) to at least ¾ of total variables and so were included.
A PCA with extraction of eigenvalues >1 through an oblique (promax) rotation with Kaiser normalization was performed on the training sample. A Kayser Meyer Olkin measure of sampling adequacy (0.844) indicated a sufficiently large sample size while Bartlett's test of sphericity (P < 0.0001) allowed rejection of the null hypothesis that the correlation matrix was an identity matrix.
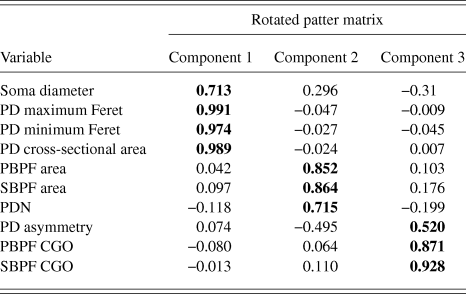
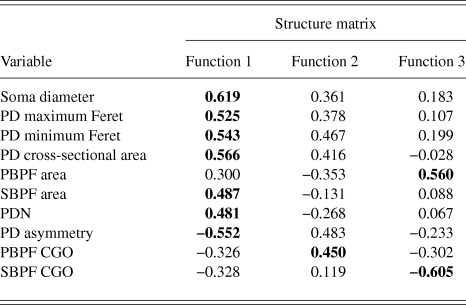
PCA produced a 3 component solution that accounted for 77.1% of the variance within the data. The rotated pattern matrix showing the variable contributions to the components is shown in Table 2. The first component comprised soma diameter, primary dendrite cross-sectional area, and minimum and maximum Feret and accounted for 45.5% of the variance in the data. This component represents the size of the RGC soma and primary dendrite thickness. The second component comprised the number of PD and PBPF and SBPF areas representing proximal dendritic tree size (accounting for 21.2% of the variance). The third component comprised both PBPF and SBPF CGO and PD range (accounting for 10.4% of the variance) thus representing asymmetry in the proximal dendritic tree. The contribution of the individual variables to the 3 dimensional separation of the RGC population is shown in Fig. 5.
Table 1. RGC morphology measurements

Values are mean ± SD.
Table 2. PCA rotated pattern matrix

Bold text denotes component to which each variable gives its greatest contribution.

Fig. 5. Principal component analysis of RGCs. PCA generated 3 components to describe variance in RGC morphology. The component scores of each RGC (n = 131) for the 3 components are plotted in X, Y, and Z dimensions. When labeled according to type (RGCA = blue, RGCB = green, RGCC = orange, RGCD = purple), this separation of the data corresponds to the 4 RGC types (A). The variable weightings toward each component are plotted in this rotated space (B). The components represent condensed variables derived from correlated or covarying variables (those encompassed by broken lines). Soma diameter, PD cross-sectional area, and minimum and maximum Feret length weight highly on component 1. Primary and SBPF area and PD number weight highly on component 2 while both the PBPF and SBPF center of gravity offset and PD asymmetry weight highly on component 3. The overlap in RGC size can be seen in the distribution of types along the first and second components (C) however taken together RGC types can be fairly separated based on the soma and proximal dendritic field size where RGCA > RGCC > RGCD > RGCB. Inclusion of the third component allows for a separation of RGC types according to asymmetry in the proximal dendritic field as evidenced by clear separation of RGCB and RGCD from RGCA and RGCC along the third component (D and E).
Proximal dendritic measurements can define RGC types
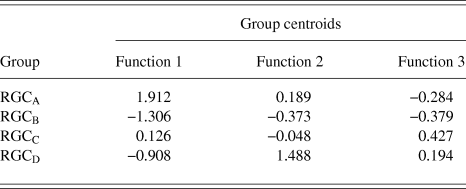
RGCs in the training sample (n = 101) were grouped using discriminant analysis. RGCs were grouped according to Sun et al. (Reference Sun, Li and He2002a). Prior probability for each RGC group was calculated based on group percentages of the training sample population to account for frequency of RGC types among a population. Discriminant analysis generated 3 discriminant functions for the separation of RGCs into groups. A chi-square statistical test of the discriminant functions showed that function 3 had no discriminating ability (P > 0.05) and so does not contribute to the separation of RGC groups. The loadings of the individual variables onto the discriminant functions are shown in Table 3. The first discriminant function accounted for 72% of between group variance within the data and describes RGC proximal dendritic tree size and symmetry, soma size and dendrite thickness. It correlated positively with soma diameter, primary dendrite cross-sectional area, minimum and maximum Feret, SBPF area, and number of PDs. The first discriminant function also showed a negative correlation with PD range. The second discriminant function (which accounted for 22% of the remaining between group variance) correlated positively with PBPF CGO, describing asymmetry in the proximal dendritic field, with a contribution from soma and dendrite thickness. The third discriminant function, accounting for the remaining 6% of between group variance; the function showed a negative correlation to SBPF CGO and a positive correlation to PBPF area. The largest determinant of RGC classification is therefore primary dendrite thickness and soma and proximal dendritic tree size, with a smaller contribution of asymmetry of the proximal dendritic tree. This is evident in the clear separation of the group centroids (Fig. 4) where the first discriminant function separates all RGC types in the order of size (where RGCA > RGCC > RGCD > RGCB).
Table 3. Correlations between variables and discriminant functions

Bold text denotes function to which each variable gives its greatest contribution.
Table 4. RGC group Centroid

RGC types discriminated by proximal dendritic morphology
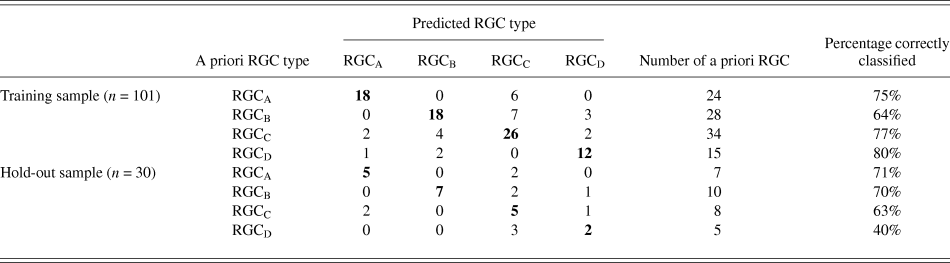
The discriminant functions correctly classified 73.3% (n = 74) of RGCs from the training sample and 63.3% (n = 19) from the hold out sample. This was greater than the correct by-chance classification of 25%. Classification is predicted based on RGC group centroids which are summarized in Table 4. The discriminant scores are plotted in Figure 6 to demonstrate the sub grouping of RGCs. The predicted group percentages for each RGC type are summarized in Table 5. RGC groups can usefully be discriminated by primary and secondary dendrite variables. Misclassification followed a similar trend with roughly 25% misclassified to the RGC type closest in morphology to the correct group. All misclassified RGCs had at least 1 variable (average 3 variables) that was >1 standard deviation from the mean of its correct type.
Table 5. Summary of classification

The relationship between population wide variance and between group variance was explored using the component scores generated through PCA as variables to describe the morphology of each RGC. The components alone achieved only a correct classification in 62.6% of the training sample (n = 101) and 60% of the hold out sample (n = 30). This indicates that population variance in morphology does not adequately describe the difference between groups.
RGC subgroups cannot be differentiated
Discriminant analysis was performed as described initially but where RGCs were grouped according to the subgroups described by Sun et al. (Reference Sun, Li and He2002a). The original hold out sample did not contain two of the subgroups and so the sample size was increased to 40 RGCs following the same random assignment method. Again, prior probability was calculated based on subgroup frequency. Correct classification was achieved in 53.8% (n = 49) of RGCs from the training sample and 35% (n = 14) from the hold out sample. While greater than the correct by chance classification (8.3%), it does not yield adequate classification indicating that the ability to resolve subgroups is beyond this model.
Discussion
Our data support the concept that RGCs in the rat retina can be classified using dendrite features that are relatively resistant to early degenerative events. It is important to note that the classification is not as precise as that provided by the dendritic tree as a whole (which includes tertiary and higher dendritic structures); however, the model provides a framework to check for labeling bias in the analysis of RGCs in retinal disease. Our data indicate that eccentricity is not a factor that biased RGC classification on the basis of soma size.
We show that RGCs can be discriminated on the basis of soma and proximal dendritic field size. This is in agreement with Sun et al. (Reference Sun, Li and He2002a) and other classifications based on RGC morphology where larger RGCA and RGCC are separated from the smaller RGCB and RGCD based on soma and dendritic field size. We also demonstrate dendrite thickness as an effective parameter for classifying RGC types due to its strong relationship with other features of dendritic tree morphology. Primary dendrite thickness has been found to correlate with soma size, dendritic field size, total dendritic length, and branching density in rat triceps surae motoneurons (Chen & Wolpaw, Reference Chen and Wolpaw1994) and cat spinal α-and γ-motoneurons (Ulfhake & Cullheim, Reference Ulfhake and Cullheim1981; Ulfhake & Kellerth, Reference Ulfhake and Kellerth1981), all parameters used in the classification of RGC types. We also show that asymmetry in the proximal dendritic tree has a small discriminatory effect on RGC types.
The use of DiOlistic labeling resulted in a number of cells being excluded from analysis as the soma and proximal dendrites were obscured by excessive labeling. However, we justify the use of DiOlistic (rather than Biolistic) labeling since this is not contingent on cell health (Honig & Hume, Reference Honig and Hume1986). Studies in the mouse glaucoma model suggest that the level of reporter expression may be affected by neuronal viability (Williams et al., Reference Williams, Howell, Barbay, Braine, Sousa, John and Morgan2013). This is vital given that the classification purpose is for use in degeneration where Biolistics is affected by the viability of transcriptional pathways necessary for the expression of fluorescent markers. Ballistic labeling techniques are open to the criticism that they bias for larger cell types since the likelihood of a hit increases with cell size. We did not observe this bias in our sample suggesting that the difference in cell soma size is insufficient to have a large effect on labeling probability. A higher proportion of RGCA (at 24%) was found in comparison to the 18% reported by Sun et al. (Reference Sun, Li and He2002a); however, this was much smaller than the 60% previously reported (Huxlin & Goodchild, Reference Huxlin and Goodchild1997). If soma size were the largest determinant of a given cell being labeled, one would expect a higher proportion of RGCA than RGCB; almost twice as many RGCB to RGCA were labeled in this study. In fact, retrograde labeling methods, such as those used by Huxlin and Goodchild (Reference Huxlin and Goodchild1997) bias toward the labeling of larger cell types, which further justifies the use of DiOlistics.
The robust discrimination between cell types is important in the study of degeneration, both in terms of reducing pseudodegenerative effects arising from intertype differences in dendritic morphology and also in investigating intertype susceptibility to disease. Type specific degeneration has been noted in the primate (Weber et al., Reference Weber, Kaufman and Hubbard1998), cat (Shou et al., Reference Shou, Liu, Wang, Zhou and Zhao2003), mouse (Leung et al., Reference Leung, Weinreb, Li, Liu, Lindsey, Choi, Liu, Cheung, Ye, Qiu, Chen, Yung, Crowston, Pu, So, Pang and Lam2011; Feng et al., Reference Feng, Zhao, Yoshida, Chen, Yang, Kim, Cang, Troy and Liu2013), and rat (Thanos, Reference Thanos1988) but not in DBA/2J (Jakobs et al., Reference Jakobs, Libby, Ben, John and Masland2005) or thy1-YFP-H transgenic mice with experimental glaucoma (Kalesnykas et al., Reference Kalesnykas, Oglesby, Zack, Cone, Steinhart, Tian, Pease and Quigley2012). The Sholl plots in Fig. 3 demonstrate the differences in branching pattern and density between RGC types and illustrate effectively how disparity in cell types between populations could be confused for atrophy in studies of degeneration.
We anticipate that the chosen parameters will remain stable during degeneration given that significant reduction in branching occurs only in the distal region of the dendritic tree, as illustrated in Fig. 1. Measurement of soma diameter is also included based on its greater resistance to degeneration than the dendritic tree. The dendritic tree exhibits the earliest change in RGCs under degeneration while the soma only undergoes substantial change with advanced/chronic glaucoma (Weber et al., Reference Weber, Kaufman and Hubbard1998). Significant change in RGC soma size in the rat is seen in chronic glaucoma models, however is similar across all retinal regions (Urcola et al., Reference Urcola, Hernandez and Vecino2006) and so is unlikely to vary between types. The ability of the system to classify cell types may therefore diminish with substantial degeneration, as soma and secondary and primary dendrite degeneration occurs. The inclusion of dendritic stratification depth within the IPL could enhance the correct classification of RGC types, in particular the classification of RGCD due to their bistratified dendritic tree.
The reproducibility of this model requires testing on a range of degenerated RGC populations, the scope of which was beyond the current study. However, applying the model to a degenerated population of RGCs would not serve to validate the model given that the correct type of an RGC cannot be determined from its post-degenerative morphology while following the degeneration of single RGCs over time is reliant on reporters whose expression is compromised in degeneration.
We would not anticipate complete neuronal classification in view of the large variability in RGC morphology and overlap between groups for any single morphological criterion. Consistent with this, all misclassified RGCs exhibited one or more variable measurement >1SD from the mean of its type. The correct subtype classification of just over a third of RGCs provides encouragement for success if given a larger population. It is also possible that the misclassified atypical cells represent previously unidentified cell types given that the number of RGC types currently defined in the closely related mouse is 22 (Voelgyi et al., Reference Voelgyi, Chheda and Bloomfield2009). The main role of this classification procedure is therefore not the definitive classification of all RGC types but to provide a degree of confidence regarding RGC proportions in a sample population where traditional methods of classification are vulnerable to bias.
Acknowledgments
The authors gratefully acknowledge Dr. Jonathan Gillard for his advice on the statistical methodology for classification.
Funding
Biotechnology and Biological Research Council grant number BB/F016352/1 and Cardiff University.