Introduction
Protoporphyrinogen oxidase (PPO) is an enzyme that catalyzes the synthesis of chlorophyll and heme in plants by converting protoporphyrinogen IX to protoporphyrin IX via oxidation (Jacobs and Jacobs Reference Jacobs and Jacobs1984). When PPO is inhibited, synthesis of these compounds is disrupted, eventually resulting in contact leaf burn (Orr and Hess Reference Orr and Hess1982; Scalla et al. Reference Scalla, Matringe, Camadro and Labbe1990). PPO is inhibited by various classes of herbicides that have been used for more than 50 yr to control weeds in agronomic crops (Dayan et al. Reference Dayan, Owens, Tranel, Preston and Duke2014). PPX1 and PPX2 are two different nuclear genes that encode two isoforms of PPO; PPO1 is targeted to plastids, and PPO2 is targeted to mitochondria (Lermontova et al. Reference Lermontova, Kruse, Mock and Grimm1997). In at least some species, the enzyme encoded by PPX2 is dual targeted to both plastids and mitochondria (Dayan et al. Reference Dayan, Barker and Tranel2018). Thus, a mutation in this gene can result in target-site resistance in both organelles (Patzoldt et al. Reference Patzoldt, Hager, McCormick and Tranel2006).
Common waterhemp [Amaranthus tuberculatus (Moq.) Sauer var. rudis (Sauer) Costea and Tardif] with resistance to PPO-inhibiting herbicides was observed in 2001 (Shoup et al. Reference Shoup, Al-Khatib and Peterson2003), and it was the first weed reported to have evolved resistance to these herbicides (Heap Reference Heap2019). The mechanism for this resistance was found to be a deletion of the amino acid glycine at the 210th position (ΔGly-210) of PPO2 (Patzoldt et al. Reference Patzoldt, Hager, McCormick and Tranel2006). The region of the sequence spanning position 210 contains two overlapping trinucleotide repeats; the loss of one repeat unit via a proposed DNA polymerase slippage-like mechanism results in the loss of a glycine codon (Gressel and Levy Reference Gressel and Levy2006). The loss of this amino acid alters the enzyme’s binding site without hindering substrate affinity but reduces sensitivity to some PPO-inhibiting herbicides by at least 100-fold (Dayan et al. Reference Dayan, Daga, Duke, Lee, Tranel and Doerksen2010). Since resistance to PPO inhibitors was reported in A. tuberculatus, 12 other weed species have evolved resistance to these herbicides (Heap Reference Heap2019).
Palmer amaranth (Amaranthus palmeri S. Watson) is one of the species that subsequently evolved resistance to PPO inhibitors. Because of widespread loss of A. palmeri control with glyphosate due to glyphosate resistance across the United States, growers have increasingly relied on PPO-inhibiting herbicides and, consequently, concerns have been raised over selection for A. palmeri resistant to PPO inhibitors (Whitaker et al. Reference Whitaker, York, Jordan and Culpepper2010).
A study by Riggins and Tranel (Reference Riggins and Tranel2012) found that A. palmeri has the same repeat motif in PPX2 as does A. tuberculatus, which suggested that A. palmeri could evolve resistance to PPO-inhibiting herbicides via the same Gly-210 deletion. This was confirmed when A. palmeri was documented as having evolved resistance to PPO inhibitors via the ΔGly-210 mutation (Salas et al. Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016). Subsequently, Giacomini et al. (Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) discovered two new mutations in PPX2 in A. palmeri that confer resistance to PPO inhibitors. These mutations confer an amino acid change at position 128, resulting in an arginine to glycine or an arginine to methionine (Arg-128-Gly and Arg-128-Met) substitution. Giacomini et al. (Reference Giacomini, Umphres, Nie, Mueller, Steckel, Young, Scott and Tranel2017) stated that the Arg-128-Gly mutation could also occur in A. tuberculatus as a result of a single nucleotide change. More recently, non–target site resistance has been confirmed and characterized in A. palmeri. Varanasi et al. (Reference Varanasi, Brabham and Norsworthy2018a) found a population of A. palmeri that was resistant to the PPO inhibitor fomesafen, likely via cytochrome P450 and gluthathione S-transferase activity.
Herbicide resistance occurs via evolution, in which the frequency of resistance alleles (and, therefore, resistant plants) in a population increases over time in response to recurrent herbicide selection (Jasieniuk et al. Reference Jasieniuk, Brûle-Babél and Morrison1996). The repeated occurrence of herbicide resistance in many weed species is a modern-day example of convergent/parallel evolution (Baucom Reference Baucom2016). Although the distinction between convergent and parallel evolution originally related to the phylogenetic distance between the species, new definitions consider the level of scale when determining whether parallel or convergent evolution is occurring (Pickersgill Reference Pickersgill2018). Specifically, at the gene level, parallel evolution involves changes in homologous genes, whereas convergent evolution involves changes in nonhomologous genes. Analogously, at the nucleotide level, parallel and convergent evolution can be distinguished by whether the change is due to the same or a different mutation. In the case of PPO inhibitor–resistant A. palmeri and A. tuberculatus, parallel evolution at the gene level would involve resistance occurring in both species as a result of changes in PPX2. If both species independently acquired an identical PPX2 change, the evolution could also be considered parallel at the nucleotide sequence level. In contrast, different PPX2 mutations between the two species would be considered parallel evolution at the gene level, but convergent evolution at the nucleotide sequence level.
Alternative to independent evolution of resistance, transfer of resistance can occur when weeds that are close relatives hybridize. Specifically, Nandula et al. (Reference Nandula, Wright, Bond, Ray, Eubank and Molin2014) found evidence that a hybridization event between spiny amaranth (Amaranthus spinosus L.) and A. palmeri resulted in the transfer of the 5-enolpyruvylshikimate-3-phosphate synthase amplicon from glyphosate-resistant A. palmeri to A. spinosus, resulting in A. spinosus becoming resistant to glyphosate as well. There have also been recorded instances of A. palmeri hybridizing with A. tuberculatus, resulting in the transfer of acetolactate synthase–inhibitor resistance (Franssen et al. Reference Franssen, Skinner, Al-Khatib, Horak and Kulakow2001; Wetzel et al. Reference Wetzel, Horak, Skinner and Kulakow1999) and glyphosate resistance (Gaines et al. Reference Gaines, Ward, Bukun, Preston, Leach and Westra2012). However, the rates of hybridization between these two species have been shown to be very low, and most hybrid progeny were also found to be sterile or nonviable (Franssen et al. Reference Franssen, Skinner, Al-Khatib, Horak and Kulakow2001; Steinau et al. Reference Steinau, Skinner and Steinau2003; Trucco et al. Reference Trucco, Zheng, Woodyard, Walter, Tatum, Rayburn and Tranel2007).
In a field in Kentucky in 2015, inadequate control of Amaranthus plants was observed following application of a PPO inhibitor. Morphological observations indicated survivors included both A. tuberculatus and A. palmeri. The purpose of this study was to (1) confirm species identity and PPO-inhibitor resistance, (2) determine the mechanism(s) responsible for resistance, and (3) determine whether the A. palmeri in this field hybridized with A. tuberculatus to acquire the gene with this mutation, or whether A. palmeri acquired resistance independently of A. tuberculatus via some form of convergent/parallel evolution.
Materials and Methods
Species Identification of Kentucky Amaranthus Samples
Seed was collected in the fall of 2015 from 10 suspected A. palmeri and 10 suspected A. tuberculatus plants that survived fomesafen plus S-metolachlor application in a soybean [Glycine max (L.) Merr.] field located in Ballard County, KY. Seed from each individual plant was designated as an accession and named KLPA1 through KLPA10 for A. palmeri and KLWH1 through KLWH10 for A. tuberculatus. Although morphology (Sauer Reference Sauer1955) of the progeny from these accessions matched the suspected species identification of the parent plants, species identification was molecularly verified. Seeds from arbitrarily selected accessions KLPA2, KLPA3, KLPA4, KLWH1, and KLWH2 were germinated on damp filter paper in petri dishes in a growth chamber at 35 C for 12 h under light conditions and 15 C for 12 h under dark conditions. After radicle emergence, the seeds were transplanted into Cone-tainers® (Stuewe and Sons, Tangent, OR, USA) containing a mixture of Sunshine® LC1(Sun Gro Horticulture, Agawam, MA, USA), soil, peat, and torpedo sand (3:1:1:1 by wt) plus 13-13-13 Osmocote® (Scotts Company, Marysville, OH, USA). The plants were grown in a greenhouse maintained at 30/25 C day/night in ambient humidity with artificial lighting from metal-halide lamps (600 μmol photon m−2 s− 1) programmed for a 16-h photoperiod. When seedlings reached the 4- to 6-leaf stage, tissue samples were collected from the most recently unfolded leaf, and DNA was extracted from the samples using the hexadecyltrimethyl-ammonium bromide (CTAB) method (Doyle and Doyle Reference Doyle and Doyle1990). A quantitative PCR (qPCR) assay (Schmittgen and Livak Reference Schmittgen and Livak2008) was performed on a subset of four samples each of suspected A. tuberculatus (two samples each from KLWH1 and KLWH2) and A. palmeri (two samples from KLPA2, and one each from KLPA3 and KLPA4) DNA from the Kentucky progeny, along with three controls each of known A. tuberculatus and A. palmeri DNA. Primers for both A. tuberculatus and A. palmeri identification, based on the ribosomal internal transcribed spacer (ITS), were used as previously described (Murphy et al. Reference Murphy, Plewa, Phillippi, Bissonnette and Tranel2017; Murphy and Tranel Reference Murphy and Tranel2018). CPS, a previously identified single-copy control gene that encodes the large subunit of carbamoylphosphate synthetase, was included (Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013). Each qPCR reaction contained approximately 50 ng DNA, 1X concentration of iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), and 0.5 μM of the forward and reverse primers. Samples were analyzed with each of the three primer sets separately and with three technical replicates.
Confirmation of PPO-Inhibitor Resistance in Kentucky Amaranthus palmeri and Amaranthus tuberculatus Populations
An initial spray study was conducted to validate field observations that the Kentucky populations were resistant to a PPO inhibitor. An even (by weight) mixture of seeds from all 10 A. tuberculatus accessions and another even mixture of seeds from 9 (seed of one accession was no longer available) of the A. palmeri accessions were first created. Sensitive (KLPAS, WHWT) accessions were also used as controls. KLPAS was collected in 2013 from a PPO inhibitor–sensitive A. palmeri plant in Fulton County, KY. WHWT comprised multiple wild-type A. tuberculatus accessions collected from various counties in Illinois in 2003. Approximately 100 seeds from each of the four groups were broadcast into 12 cm by 12 cm greenhouse flat inserts containing a mixture of Sunshine® LC1, soil, peat, and torpedo sand (3:1:1:1 by wt) plus 13-13-13 Osmocote®. When the seedlings reached the 1- to 2-leaf stage, they were transplanted into 8 cm by 8 cm square plastic pots containing the same mixture of Sunshine® LC1, soil, peat, and torpedo sand plus fertilizer. Uniform, 8- to 10-cm-tall plants were arbitrarily selected for herbicide application (18 Kentucky A. palmeri plants, 15 Kentucky A. tuberculatus plants, 5 KLPAS plants, and 6 WHWT plants). Tissue samples for subsequent DNA extraction using the CTAB method were collected before herbicide application. Plants were sprayed with lactofen (Cobra®, 240 g ai L−1, Valent USA, Walnut Creek, CA, USA) using a compressed-air laboratory spray chamber (DeVries Manufacturing, Hollandale, MN, USA) equipped with an 80° even flat-fan spray nozzle (TeeJet Technologies, Wheaton, IL, USA) and calibrated to deliver 187 L ha−1. Amaranthus tuberculatus plants were sprayed at a rate of 13.7 g ai ha−1, and A. palmeri plants were sprayed at a rate of 110 g ha−1; these rates were determined in preliminary studies to distinguish resistant and sensitive plants for each of the two species. All spray mixtures contained 1% v/v crop oil concentrate (COC). At 2 wk after treatment (WAT), plants were classified as resistant if they exhibited healthy new growth.
Genotyping of Kentucky Amaranthus palmeri and Amaranthus tuberculatus Plants
A qPCR TaqMan assay was performed on the DNA of resistant plants from the spray study to determine the presence or absence of the ΔGly-210 mutation. This allele-specific assay uses fluorescent probes designed by Wuerffel et al. (Reference Wuerffel, Young, Lee, Tranel, Lightfoot and Young2015) to detect the ΔGly-210 mutation. The TaqMan assay was conducted using the allelic discrimination program in the Applied Biosystems 7900 HT fast real-time PCR system (Applied Biosystems, Foster City, CA, USA) at the W.M. Keck Center at the Roy J. Carver Biotechnology Center at the University of Illinois, Urbana–Champaign.
Sequencing of the Gly-210 Region of Kentucky Amaranthus palmeri and Amaranthus tuberculatus Samples
Once it was found that the ΔGly-210 mutation was present in both the A. tuberculatus and the A. palmeri populations, samples of each species that were homozygous (based on the TaqMan assay) for the deletion and samples that were homozygous wild type were sequenced. To sequence the selected samples, the primers PPX2Lex7F1 (5′-ATTATGTTGGAACCATTTCT) and PPX2Lex9R1 (5′-GATAGCGATTGAGGATCT) were used to amplify a 700-bp region spanning exon 7 to exon 9 of PPX2, which included the deletion site. Amplification reactions totaled 25 μl and contained approximately 100 ng DNA, 1 U Taq polymerase (New England Biolabs, Ipswich, MA, USA), 1.0 mM MgCl2, 0.2 mM each deoxyribonucleotide triphosphate, and 0.1 μM of the forward and reverse primers. The PCR protocol was as follows: incubation for 5 min at 95 C followed by 34 cycles of 0.5 min at 95 C, 0.5 min at 54 C, and 2 min at 72 C, with a final extension step of 2 min at 72 C. A portion of the PCR product was then fractionated on a 1% agarose gel plus 0.4% GreenGlo™ Safe DNA Dye (Denville Scientific, Metuchen, NJ, USA) at 120 V. PCR products were visualized using ultraviolet light to confirm that amplification of the correct product occurred. Following agarose gel fractionation, the bands were excised, and the DNA was purified from the gel using a QIAEX II Gel Extraction Kit (Qiagen, Germantown, MD, USA). The purified product was sequenced using an AB BigDye Terminator v 3.1 Cycle Sequencing Kit (Applied Biosystems) using the forward primer, PPX2Lex7F1. The sequenced products were then analyzed by the W.M. Keck Center with an ABI 3730xl Capillary DNA Analyzer (Applied Biosystems). The resulting sequence data were analyzed using Sequencher v. 5.4 software (Gene Codes, Ann Arbor, MI, USA). Sequences were aligned using MAFFT v. 7 (Katoh et al. Reference Katoh, Misawa, Kuma and Miyata2002).
Phylogenetic Analysis of the Gly-210 Region of Kentucky Amaranthus palmeri and Amaranthus tuberculatus Samples
Aligned sequences were further analyzed using Molecular Evolutionary Genetics Analysis (MEGA v. 10) software (Kumar et al. Reference Kumar, Stecher, Li, Knyaz and Tamura2018) to investigate possible hybridization. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei Reference Saitou and Nei1987; Tamura et al. Reference Tamura, Nei and Kumar2004). Ambiguous base pairs (likely due to heterozygosity) were coded using the International Union of Pure and Applied Chemistry (IUPAC) ambiguity code (Tipton Reference Tipton1994). The Tamura three-parameter method was used to determine evolutionary distances, and a pairwise-deletion sequence comparison was done on the 1st, 2nd, 3rd, and noncoding positions. Confidence in the neighbor-joining tree was estimated by 1,000 bootstrap replicates.
Exploration of Additional Resistance Mutations in the Kentucky Amaranthus palmeri Population
Once it was determined that additional resistance mutations might be present in the Kentucky A. palmeri population, seeds from nine of the A. palmeri accessions were planted in petri dishes and transplanted into Cone-tainers® as described previously. Plants (6 to 20 per accession) were sprayed with lactofen at a rate of 5.50 g ha−1 when they reached 5 to 6 cm in height. This rate was much lower than that used in the resistance confirmation study, because the plants were sprayed at a smaller height. All spray mixtures contained 1% v/v COC. At 2 WAT, plants were classified as resistant if they exhibited healthy new growth, and tissue was collected from survivors. DNA was extracted from the tissue using the CTAB method, and a TaqMan assay was performed on the DNA to determine whether the ΔGly-210, Arg-128-Gly, and Arg-128-Met mutations were present. The TaqMan primer and probe combinations used for detection of the R128 mutations were previously reported by Varanasi et al. (Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2018b).
Results and Discussion
Species Identification of Kentucky Amaranthus Samples
The qPCR-based approach of using species-specific primers for A. tuberculatus and A. palmeri in addition to primers for the single-copy control gene, CPS, allowed for confident discrimination of A. tuberculatus and A. palmeri samples from each other (Figure 1). The cycle threshold (Ct) values for known and suspected samples of each species were tested with the primers for both A. tuberculatus and A. palmeri identification and graphed against the Ct values for the CPS primers. In both cases, the suspected A. palmeri samples grouped with the known A. palmeri samples, and the suspected A. tuberculatus samples grouped with the known A. tuberculatus samples. These results confirmed that the two species in this field were A. palmeri and A. tuberculatus. Additionally, the strong clustering of unknown A. palmeri with known A. palmeri and unknown A. tuberculatus with known A. tuberculatus was not consistent with hybridization having occurred between the two species.
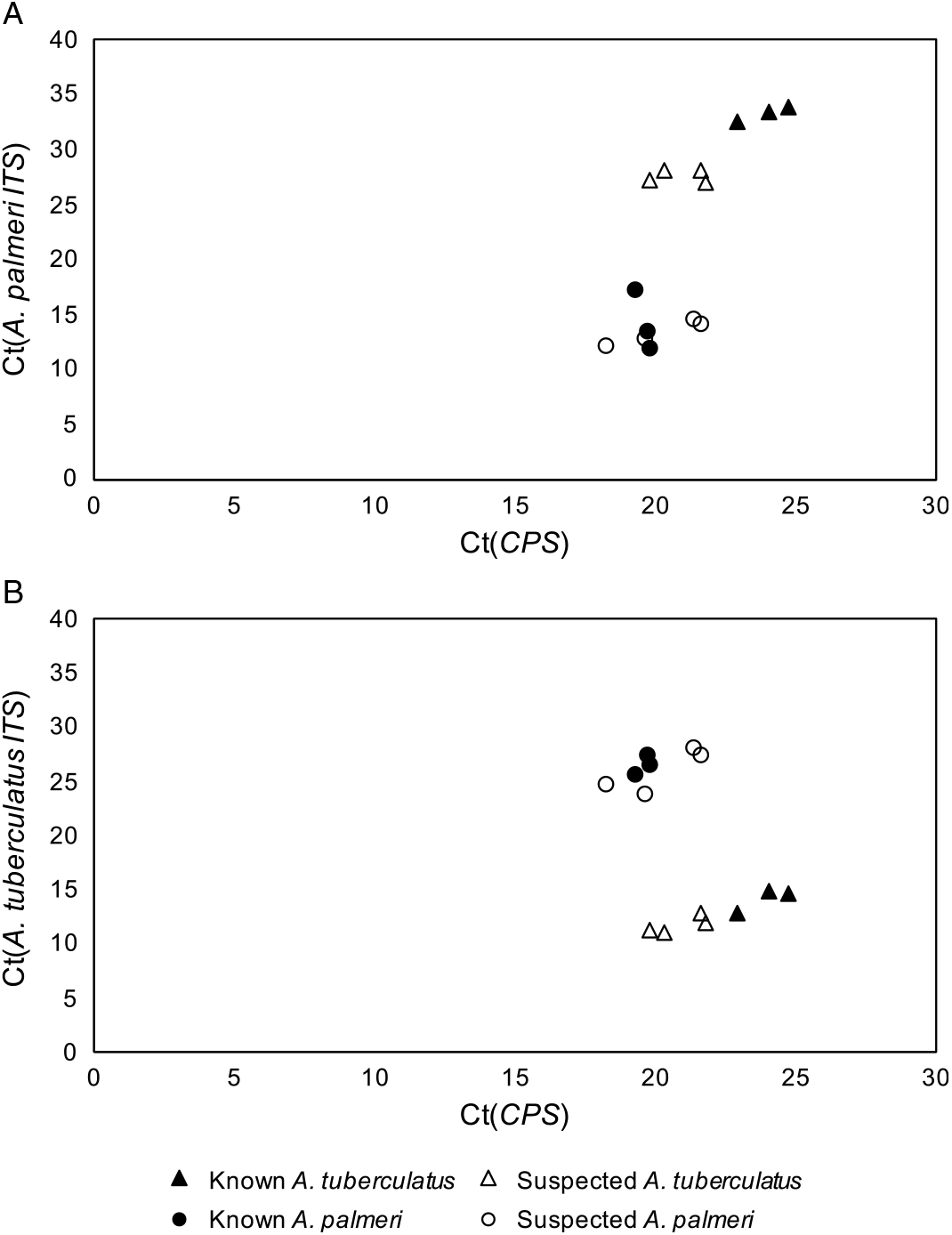
Figure 1. Quantitative polymerase chain reaction cycle threshold (Ct) values of Amaranthus tuberculatus (A) and Amaranthus palmeri (B) ribosomal internal transcribed spacer (ITS) and reference gene (CPS) across known and suspected A. palmeri and A. tuberculatus samples.
Confirmation of PPO-Inhibitor Resistance in Kentucky Amaranthus palmeri and Amaranthus tuberculatus Populations and Genotyping of Resistant Plants
The results of the spray study confirmed that the two populations in this field were resistant to lactofen. All plants from the suspected resistant populations of both species survived lactofen, whereas all plants of the sensitive control populations did not (Table 1). Genotyping of the resistant plants revealed that all A. tuberculatus plants tested contained the ΔGly-210 mutation. In contrast, only one-third of the resistant A. palmeri plants had the ΔGly-210 mutation (Table 1).
Table 1. Confirmation of lactofen resistance and percent of resistance accounted for by the ΔGly-210 mutation in Kentucky (KY) Amaranthus tuberculatus and KY Amaranthus palmeri accessions.

a Known lactofen-sensitive populations were included as controls. KLPAS, A. palmeri; WHWT, A. tuberculatus).
Phylogenetic Analysis of the Gly-210 Region of Kentucky Amaranthus palmeri and Amaranthus tuberculatus Samples
Although results from the species-specific markers suggested there was no hybridization, we sought to further verify that the ΔGly-210 mutation independently evolved in both species by conducting a phylogenetic analysis of PPX2. The sequence surrounding the Gly-210 site in exon 9 of PPX2 is highly conserved and thus does not contain many polymorphic sites (Thinglum et al. Reference Thinglum, Riggins, Davis, Bradley, Al-Khatib and Tranel2011). Inference of a phylogenetic tree is best done on highly polymorphic regions, so the region between exons 7 and 8 of PPX2 was included for analysis, along with the entirety of exon 8. After the ends of the sequences were trimmed, the sequences contained 480 bp, with 62 nucleotide polymorphisms and four deletions (located in introns) among 10 individuals (five of each species). Only 10 of the nucleotide polymorphisms were located in exons. The sum of branch lengths was calculated to be 0.130 under the neighbor-joining method. Dioecious plants can have a high level of heterozygosity, and any sequence ambiguities likely due to this heterozygosity were scored with an IUPAC ambiguity code (Tipton Reference Tipton1994). In the aligned sequences, less than 5% of the sites were ambiguous.
The A. tuberculatus and A. palmeri samples, some of which were homozygous for the ΔGly-210 mutation and some of which were homozygous wild type, grouped together by species, and they grouped with known samples of their respective species (Figure 2). This indicates that the A. palmeri and A. tuberculatus growing together in this field did not hybridize with each other to pass on resistance; rather, they evolved the ΔGly-210 mutation independently. This was somewhat expected, because both species contain the same repeat motif in PPX2, which is conducive to the evolution of this mutation (Riggins and Tranel Reference Riggins and Tranel2012) and, at the time of this project, the ΔGly-210 mutation already had been reported in A. palmeri (Salas et al. Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016). Furthermore, previous research has found that hybridization between A. palmeri and A. tuberculatus results in predominantly nonviable progeny (Franssen et al. Reference Franssen, Skinner, Al-Khatib, Horak and Kulakow2001; Steinau et al. Reference Steinau, Skinner and Steinau2003; Trucco et al. Reference Trucco, Zheng, Woodyard, Walter, Tatum, Rayburn and Tranel2007), so hybridization between the two populations in this field seemed unlikely.
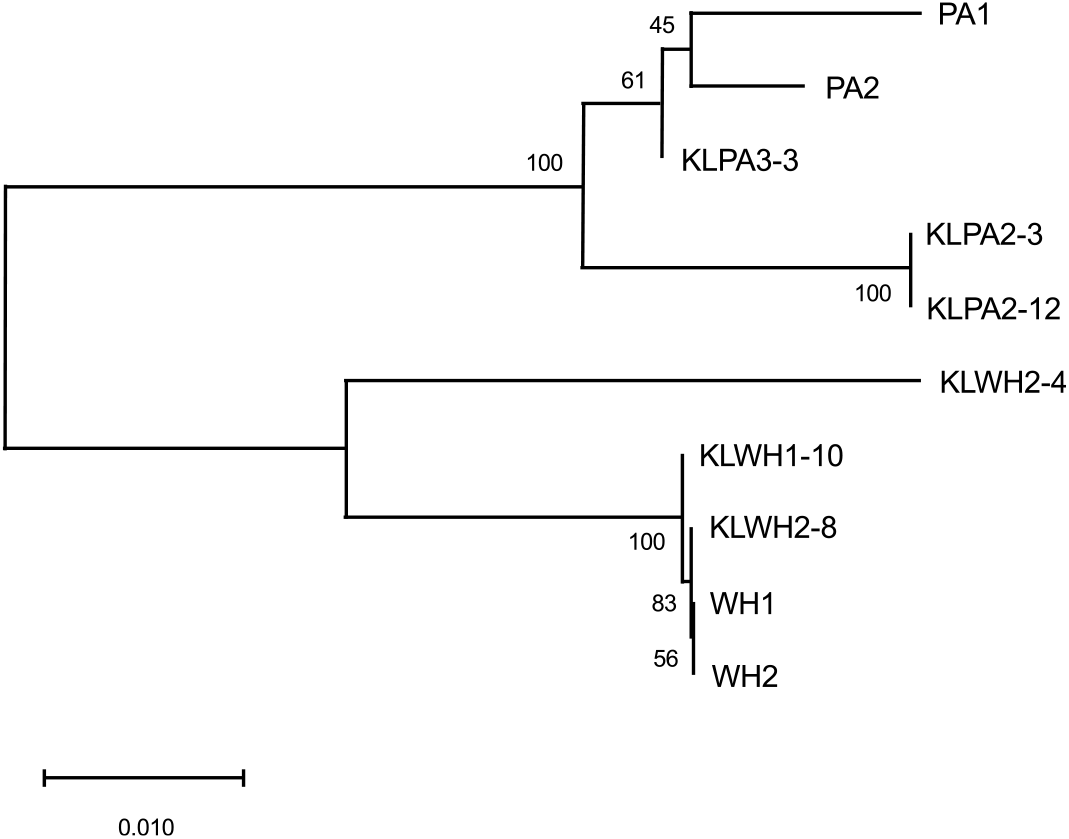
Figure 2. Neighbor-joining tree showing relationships of a 480-bp region of PPX2. Evolutionary distances were calculated using the Tamura three-parameter method and are in the units of the number of base substitutions per site. The tree is drawn to scale, with the branch lengths also in the units of number of base substitutions per site, as depicted by the scale bar. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. Each taxon on the tree represents one plant. KLPA and KLWH are plants from Amaranthus palmeri and Amaranthus tuberculatus populations, respectively, in Kentucky that exhibited protoporphyrinogen oxidase–inhibitor resistance; KLPA3-3 lacked the PPX2 mutation conferring the Gly-210 deletion, whereas the other plants were homozygous for this mutation. PA1 and PA2 and WH1 and WH2 are known samples of A. palmeri and A. tuberculatus, respectively; WH1 was homozygous for the Gly-210 deletion mutation, whereas the other plants lacked this mutation.
Exploration of Additional Resistance Mutation in the Kentucky Amaranthus palmeri Population
During the course of this research, two additional PPX2 mutations in A. palmeri were identified, so it was suspected that the Kentucky A. palmeri population might also contain either or both of these mutations. Upon testing, it was found that the Arg-128-Gly mutation was present in all nine Kentucky A. palmeri accessions (Figure 3). The Arg-128-Met mutation was not found, which was not entirely surprising, because it is less common than the Arg-128-Gly mutation (Copeland et al. Reference Copeland, Giacomini, Tranel, Montgomery and Steckel2018; Varanasi et al. Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2018b). The percent of resistant plants in these accessions ranged from 24% in accessions KLPA7 and KLPA9 to 88% in accession KLPA2. The percent of resistant plants in these accessions containing only the Arg-128-Gly mutation ranged from 11% in accession KLPA8 to 80% in accession KLPA4. The percent of resistant plants containing only the ΔGly-210 mutation ranged from 0% in accession KLPA4 to 83% in accession KLPA8. In six of the nine accessions, there were resistant plants that contained both the Arg-128-Gly and the ΔGly-210 mutations. However, these two mutations might not have occurred on the same allele, because all plants containing both mutations were heterozygous for each, based on the TaqMan assay (data not shown). Overall, 51% of resistant plants among all tested accessions had the ΔGly-210 mutation only, 30% had only the Arg-128-Gly mutation, and 19% carried both the ΔGly-210 and the Arg-128-Gly mutations. All resistant plants had one or both of the mutations, and none of the sensitive plants had either of the mutations.
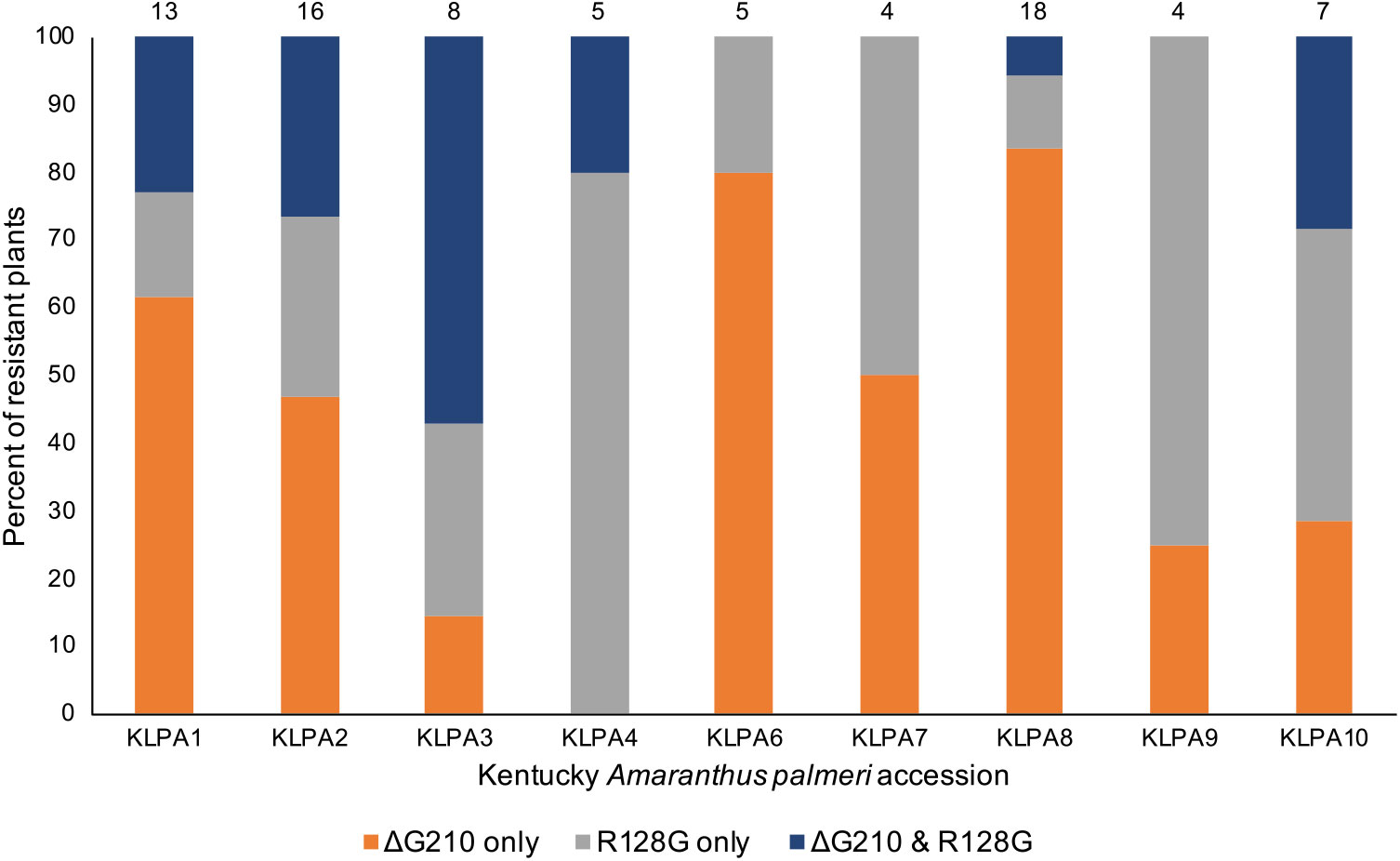
Figure 3. Percent of resistant plants containing each PPX2 resistance mutation or combination of mutations in each Kentucky Amaranthus palmeri accession. The number of resistant plants evaluated from each accession is shown above each bar.
Although a detailed history is unavailable for the field in which the two populations used in this study were collected, soybean was planted in 2015 and in at least the four prior years, and there were multiple applications of PPO-inhibiting herbicides during this time. Why A. palmeri evolved two mechanisms of resistance and A. tuberculatus only one in this field is unclear. The Arg-128-Gly mutation should be just as likely to evolve in A. tuberculatus, as it would take one nucleotide change within the wild-type A. tuberculatus arginine codon to result in a substitution to glycine. Perhaps the accumulation of two different mutations in the A. palmeri population was due to a combination of selection on existing genetic variation and immigration from another field. Personal communication with the producer revealed that he suspected Amaranthus spp. were introduced into the field from harvest equipment.
It should also be noted that, although the mutations conferring the Gly-210 deletion and the Arg-128-Gly substitution collectively accounted for resistance in every plant analyzed from both species, it is possible that other mechanisms of resistance to PPO inhibitors were present. Clearly there are multiple mechanisms for resistance to PPO inhibitors, and more research is needed to fully understand the relative fitness advantages and disadvantages of various resistance mechanisms to PPO inhibitors in the presence and absence of diverse PPO-inhibiting herbicides.
This study highlights the adaptability of these two species when under selection pressure from PPO-inhibiting herbicides. Resistance in both species arose independently via the ΔGly-210 mutation, rather than through transfer of resistance via hybridization, and A. palmeri further evolved an additional resistance mutation. The same evolutionary solution arose in both species—and yet another adaptation occurred in one species—to overcome an abiotic stress, providing a modern-day example of both parallel and convergent evolution at the nucleotide sequence level occurring within a single field.
Acknowledgments
The USDA National Institute of Food and Agriculture (Hatch Project ILLU-802-944) provided partial funding of this research. We thank James Martin for original collection of the Kentucky seed accessions used in this project. No conflicts of interest have been declared.





