Weed resistance to herbicides may be conferred through two major mechanisms: target and nontarget site (Powles and Yu Reference Powles and Yu2010). Target-site herbicide resistance is endowed by changes to the nucleotide sequence of the target-site gene causing missense mutation(s), leading to changes in the three-dimensional structure of the protein and hence herbicide affinity (Dayan etal. Reference Dayan, Daga, Duke, Lee, Tranel and Doerksen2010; Tranel and Wright Reference Tranel and Wright2002). Gene amplification is another commonly reported target-site mechanism in herbicide-resistant plants (Gaines etal. Reference Gaines, Zhang, Wang, Bukun, Chisholm, Shaner, Nissen, Patzoldt, Tranel, Culpepper, Grey, Webster, Vencill, Sammons, Jiang, Preston, Leach and Westra2010). In contrast, the more complex and less understood nontarget site resistance (NTSR) can be attributed to differences in herbicide uptake and translocation, metabolic detoxification, or sequestration mechanisms (Powles and Yu Reference Powles and Yu2010; Yuan etal. Reference Yuan, Tranel and Stewart2007). It was only in the past decade that the significance and implications of NTSR have started to unfold.
Evolution of NTSR is facilitated by the gradual accumulation of resistant alleles within the individuals of a population, resulting in favorable allelic combinations over time (Délye etal. Reference Délye, Jasieniuk and Le Corre2013). Outcrossing species are known to evolve NTSR mechanisms much faster than self-pollinating species (Délye etal. Reference Délye, Jasieniuk and Le Corre2013). NTSR is considered a threat to sustainability of herbicide use for chemical weed control, mainly due to its unintended consequences such as resistance to herbicides from different site-of-action (SOA) groups (Délye etal. Reference Délye, Jasieniuk and Le Corre2013; Petit etal. Reference Petit, Duhieu, Boucansaud and Délye2010). Previous reports have described NTSR mechanisms (mainly metabolic) in weeds for several SOAs, including acetyl coenzyme A carboxylase and inhibitors of photosystem II (PSII), 4-hydroxyphenylpyruvate dioxygenase (HPPD), 5-enolpyruvylshikimate-3-phosphate synthase, acetolactate synthase (ALS), and protoporphyrinogen oxidase (PPO) (Guo etal. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015; Han etal. Reference Han, Yu, Owen, Cawthray and Powles2016; Kaundun etal. Reference Kaundun, Hutchings, Dale, Howell, Morris, Kramer, Shivrain and Mcindoe2017; Ma etal. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013; Nakka etal. Reference Nakka, Godar, Thompson, Peterson and Jugulam2017; Preston and Wakelin Reference Preston and Wakelin2008; Varanasi etal. Reference Varanasi, Brabham and Norsworthy2018b; Yu etal. Reference Yu, Abdallah, Han, Owen and Powles2009).
Palmer amaranth is one of the most problematic weeds in North America, especially in midsouthern United States, due to its rapid growth rate, prolific seed production, and propensity to evolve resistance to several herbicide SOAs (Heap Reference Heap2018; Korres and Norsworthy Reference Korres and Norsworthy2017; Van Wychen Reference Van Wychen2016). PPO inhibitors became a popular option for controlling ALS-inhibitor and glyphosate-resistant Palmer amaranth biotypes in soybean and cotton (Gossypium hirsutum L.) (Owen and Zelaya Reference Owen and Zelaya2005; Rousonelos etal. Reference Rousonelos, Lee, Moreira, VanGessel and Tranel2012). However, repeated use of PPO-inhibiting herbicides for controlling Amaranthus spp. has resulted in the evolution of resistance to this SOA. To date, mostly target-site alterations have been reported to confer resistance to PPO inhibitors. The first target-site mechanism (namely, ΔG210) conferring PPO-inhibitor resistance was reported in waterhemp [A. tuberculatus (Moq.) Sauer] in Illinois (Lee etal. Reference Lee, Hager and Tranel2008; Patzoldt etal. Reference Patzoldt, Hager, McCormick and Tranel2006), followed by Palmer amaranth in Arkansas (Salas etal. Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016). Several other target-site mutations in the PPO enzyme of Palmer amaranth endowing PPO-inhibitor resistance have been confirmed: arginine to glycine/methionine (R128G/M), glycine to alanine (G399A), glycine to glutamic acid (G114E), and serine to isoleucine (S149I) (Giacomini etal. Reference Giacomini, Umphres-Lopez, Nie, Mueller, Steckel, Young and Tranel2017; Rangani etal. Reference Rangani, Salas, Aponte, Landes and Roma-Burgos2018).
An alternative resistance mechanism to PPO inhibitors became more evident when Obenland etal. (Reference Obenland, Ma, O’Brien, Lygin and Riechers2017) reported an NTSR mechanism to carfentrazone-ethyl in waterhemp. Varanasi etal. (Reference Varanasi, Brabham and Norsworthy2018b), however, confirmed and characterized the first documented case of NTSR to fomesafen in a Palmer amaranth accession from Randolph County, AR (RCA). Additional research on the cross-resistance patterns of the RCA accession was needed to understand the implications of NTSR. In light of the previous findings, the objectives of this study were to (1) determine the level of fomesafen resistance in the NTSR Palmer amaranth accession with/without malathion, (2) characterize cross-resistance patterns of the Palmer amaranth accession for different PPO class herbicides, and (3) determine the effects of malathion with other PPO-inhibiting herbicides (namely, flumioxazin, saflufenacil, and acifluorfen) in the Palmer amaranth accession.
Materials and Methods
Plant Materials
The Palmer amaranth accession under investigation was collected in 2016 from a cotton and soybean field in RCA (36° 29’ 10”N, 90° 55’ 37.92”W), as a part of a late-season survey to assess the severity of fomesafen resistance in the state (Varanasi etal. Reference Varanasi, Brabham, Norsworthy, Nie, Young, Houston, Barber and Scott2018a). The initial seed lot for the RCA accession was created by threshing 10 inflorescences. A homogeneous F1 population of the RCA accession was later created after one generation of selection and used for this study (Varanasi etal. Reference Varanasi, Brabham and Norsworthy2018b). Two known susceptible biotypes from 2007 (S1) and 1986 (S2) were included for comparison in the dose-response and cross-resistance assays.
Greenhouse experiments were conducted in 2018 at the Altheimer Laboratory, University of Arkansas, Fayetteville, AR. Palmer amaranth seed (RCA accession, S1, and S2) were germinated in plastic trays (25 cm × 55 cm), and 1 week after germination, seedlings at the one- to two-leaf stage were transplanted into 50-well plastic trays (25 cm × 55 cm) filled with potting mix (Sunshine premix No. 1®; Sun Gro Horticulture, Bellevue, WA). Soybean (var. CZ 4748LL, Bayer Crop Science) seeds were also planted in plastic pots (8-cm height × 10-cm diameter) filled with potting mix. Initially, four soybean seeds were planted in each pot, which were later thinned to one plant at the seedling stage. All plants were grown under a 16-h photoperiod, 35/25 C day/night temperature, and watered daily throughout the study.
Fomesafen (with or without Malathion) Dose-Response Experiment
To determine the level of resistance in the RCA accession, seedlings at the four- to six-leaf stage were treated with increasing rates of fomesafen with or without malathion. The following fomesafen rate structures were used for the RCA accession and known susceptible biotypes (S1 and S2) based on the 1× rate of fomesafen at 395 g ai ha−1: 0.06×, 0.12×, 0.25×, 0.5×, 1×, 2×, 4×, and 8× for the RCA accession and 0.003×, 0.007×, 0.015×, 0.03×, 0.06×, 0.12×, 0.25×, and 0.5×, for S1 and S2. A nonionic surfactant was included at 0.25% v/v in all treatments. Plants were sprayed with malathion (Hi-Yield® Malathion; Hi-Yield Chemical Company, Bonham, TX) at 1,500 g ai ha−1 2 h before fomesafen treatments (Oliveira etal. Reference Oliveira, Gaines, Dayan, Patterson, Jhala and Knezevic2018). Treatments were applied using a research track sprayer equipped with two flat-fan spray nozzles (TeeJet spray nozzles; Spraying Systems Co., Wheaton, IL) calibrated to deliver 187 L ha−1 of herbicide solution at 269 kPa, moving 1.6 km h−1.
Herbicide Cross- and Multiple Resistance Studies
To determine the cross- and multiple resistance profile of the RCA accession, seedlings at four- to six-leaf stage were treated with field recommended rates of the herbicides listed in Table 1. A known S2 biotype was included as a check and treated with the aforementioned herbicides for comparison. All herbicide treatments included label-recommended adjuvants: crop oil concentrate (Superb® HC; Winfield Solutions, St. Paul, MN) at 1% v/v for flumioxazin, saflufenacil, and atrazine; nonionic surfactant (Induce®; Helena Agri-Enterprises, Collierville, TN) at 0.25% v/v for acifluorfen, pyrithiobac sodium, and dicamba; and methylated seed oil (Loveland Products Inc., Greeley, CO) at 1% v/v for tembotrione.
Table 1. Herbicide treatments (1×) applied for testing the cross- and multiple-resistance profile of Palmer amaranth accession from Randolph County, AR.

Treatment of RCA Accession with Malathion and PPO Inhibitors
To investigate the effects of malathion with PPO-inhibiting herbicides, seedlings (four- to six-leaf stage) of the RCA accession were treated separately with flumioxazin, saflufenacil, and acifluorfen (Table 1) with or without malathion (1,500 g ai ha−1). Malathion was applied 2 h before the herbicide treatments. All PPO inhibitor treatments included label-recommended adjuvants as previously discussed.
Treatment of Soybean with Fomesafen, P450, and Glutathione S-Transferase Inhibitors
Soybean seedlings (V2) were treated separately with fomesafen (395 g ai ha−1), P450 inhibitor malathion (1,500 g ai ha−1), glutathione S-transferase (GST) inhibitor 4-chloro-7-nitrobenzofurazan (NBD-Cl; Sigma-Aldrich Ltd., St. Louis, MO) (270 g ai ha−1), malathion followed by (fb) fomesafen, or NBD-Cl fb fomesafen under greenhouse conditions (16-h photoperiod, 35/25 C day/night). The NBD-Cl rate of 270 g ai ha−1 was chosen on the basis of a study conducted in atrazine-resistant waterhemp (Ma etal. Reference Ma, Evans and Riechers2016). Malathion was applied 2 h and NBD-Cl 2 d before fomesafen treatment.
Genomic DNA Isolation
Young leaf tissue was collected from the Palmer amaranth survivors 2 wk after treatment (WAT) with pyrithiobac sodium. Tissue from seven RCA and two S plants were collected, placed in liquid nitrogen, and stored at −80 C until further use. Genomic DNA (gDNA) was isolated from the leaves using a modified cetyltrimethylammonium bromide protocol (Doyle and Doyle Reference Doyle and Doyle1987). The quantity and quality of isolated gDNA were determined using a spectrophotometer (NanoDrop 1000; Thermo Fisher Scientific, Waltham, MA) and agarose gel electrophoresis.
ALS Gene-Specific Primer Designing and Sequencing
To amplify and sequence the ALS gene (approximately 2,000 bp) from the RCA and S accessions, the following primers were used: forward 5’- CTGCAATCATCCATTTACGCTATC -3’ and reverse 5’- TCCAACCAACTAATAAGCCCTTC -3’. Primers were designed on the basis of the sequence information from Amaranthus spp. available at GenBank (accession U55852.1), using OligoAnalyzer v. 3.1 (Integrated DNA Technologies, Coralville, IA) (Owczarzy etal. Reference Owczarzy, Tataurov, Wu, Manthey, McQuisten, Almabrazi, Pedersen, Lin, Garretson, McEntaggart, Sailor, Dawson and Peek2008) and checked for specificity using the BLAST tool at the National Center for Biotechnology Information (Bethesda, MD).
Polymerase chain reactions (PCRs) were performed in a T100 thermal cycler (Bio-Rad, Hercules, CA) for amplification of the ALS gene. A 50-μl reaction volume consisted of 10 μl of 5× GoTaq Flexi buffer (Promega, Madison, WI), 4 μl of 25 mM MgCl2 (Promega), 1 μl of 10 mM dNTP mix (Promega), 0.25 μl of GoTaq Flexi DNA Polymerase (Promega), 1 μl each of forward and reverse gene-specific primers, 2 μl of gDNA, and 30.75 μl of nuclease-free water (Thermo Fisher Scientific). PCR conditions for amplifying the ALS gene were 95.0 C for 3 min, 35 cycles of 95.0 C for 30 s, 53.0 C for 30 s, and 72.0 C for 2 min, fb 72.0 C for 7 min.
The PCR products were run on 0.8% agarose gel with a 1-kb marker to confirm amplicon size, purified using the GeneJet PCR purification kit (Thermo Fisher Scientific), and quantified with a NanoDrop spectrophotometer. The purified PCR products (25 to 50 ng μl−1) were sent for Sanger DNA sequencing (Genewiz, South Plainfield, NJ), and aligned using MultAlin to identify point mutations (Corpet Reference Corpet1988).
Statistical Analysis
Experiments were conducted in two separate runs and three replications (10 plants per replication). Data on percent survivors and dry biomass (oven-dried at 65 C for 3 d) were collected 2 WAT. Dry biomass data were expressed as a percentage of the nontreated plants. The soybean experiment was carried out in three separate runs, each run consisting of six replications (one plant per replication). Aboveground fresh soybean biomass was collected at 2 WAT and converted into percent biomass reduction relative to the nontreated plants. ANOVA was performed, and data were analyzed using the JMP Pro 13.1 (SAS Institute Inc., Cary, NC).
Dose-response data were analyzed using the drc package in R 3.1.2 (R Development Core Team 2018). The relationship between herbicide rate and aboveground dry biomass was established using a four-parameter log-logistic model (Seefeldt etal. Reference Seefeldt, Jensen and Fuerst1995), described as follows:
where Y is the response (aboveground dry biomass) expressed as a percentage of the nontreated control, C is the lower limit of Y, D is the upper limit of Y, b is the slope of the curve around the effective herbicide dose for 50% biomass reduction (GR50), and x is the herbicide dose. The resistance index (resistance divided by susceptibility [R/S]) was calculated using GR50 values. There was no significant interaction between runs; therefore, data were pooled (P > 0.05).
Results and Discussion
Level of PPO-Inhibitor Resistance (with or without Malathion)
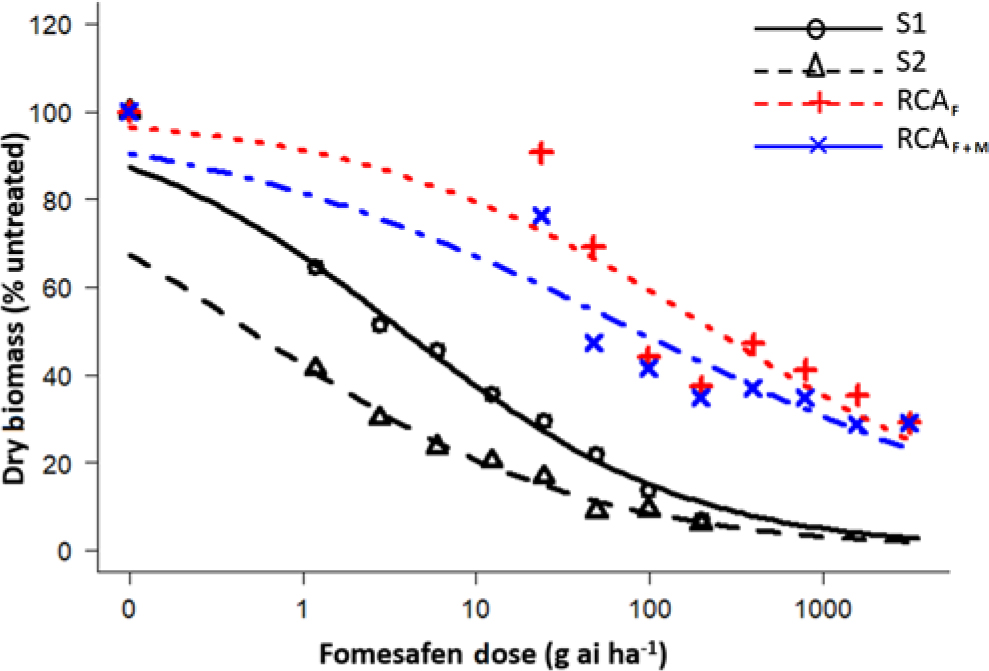
Figure 1. Dose-response assay using a four-parameter log logistic model for fomesafen-resistant Palmer amaranth Randolph County, AR (RCA), and susceptible accessions. S1, susceptible accession from 2007; S2, susceptible accession from 1986; RCAF, Randolph County, AR, treated only with fomesafen; RCAF+M, Randolph County, AR, accession treated fomesafen plus malathion.
The fomesafen doses that caused 50% reduction in the aboveground dry biomass of the S1 (2007) and S2 (1986) were 3.78 and 0.50 g ai ha−1, respectively, whereas for the RCA accession, those doses were 238 g ai ha−1 (fomesafen only [RCAF]) and 83.8 g ai ha−1 (fomesafen plus malathion [RCAF+M]) (Table 2). Based on GR50, the presence of malathion increased the efficacy of fomesafen by approximately 64%. There was a downward shift in the fomesafen dose-response curve for RCA accession in the presence of malathion (Figure 1). Varanasi etal. (Reference Varanasi, Brabham and Norsworthy2018b) had earlier reported partial reversal of fomesafen resistance in the RCA accession after application of malathion preceding fomesafen treatment at 263 g ai ha−1. Based on the GR50 values, the resistant indices (RCAF/S1 and RCAF+M/S1) for fomesafen were estimated to be 63- and 22-fold, respectively. In contrast, the fomesafen resistance indices relative to the S2 accession (RCAF/S2 and RCAF+M/S2) were 476- and 167-fold, respectively. Previously, the resistant index for the RCA accession based on the S2 accession was reported to be 18-fold (Varanasi etal. Reference Varanasi, Brabham and Norsworthy2018b). The higher level of the fomesafen resistance observed in the present study is due to the use of a more homogeneous F1 population instead of a field-derived population for the initial dose-response experiment. Studies conducted with other SOA herbicides have shown an increase in resistance levels with repeated selection in Palmer amaranth (Norsworthy Reference Norsworthy2014; Tehranchian etal. Reference Tehranchian, Norsworthy, Powles, Bararpour, Bagavathiannan, Barber and Scott2017; Varanasi etal. Reference Varanasi, Brabham, Bagavathiannan and Norsworthy2018c).
Table 2. Summary parameters describing the response of aboveground dry biomass from resistant (Randolph County, Arkansas,) and susceptible (S1 and S2) Palmer amaranth accessions to increasing rates of fomesafen 2 WAT.

a The greenhouse experiment was conducted at the Altheimer Laboratory, University of Arkansas, Fayetteville, AR.
b Abbreviations: RCA, Randolph County, AR; RCAF, Randolph County, AR, accession treated with fomesafen alone; RCAF+M, Randolph County, AR, accession treated with fomesafen plus malathion; S1, known susceptible from 2007; S2, known susceptible from 1986; GR50, 50% biomass reduction; WAT, weeks after treatment.
c GR50 values of RCAF and RCAF+M were highly significant compared with the S1 and S2 biotypes (P < 0.001).
*** P-value of < 0.001.
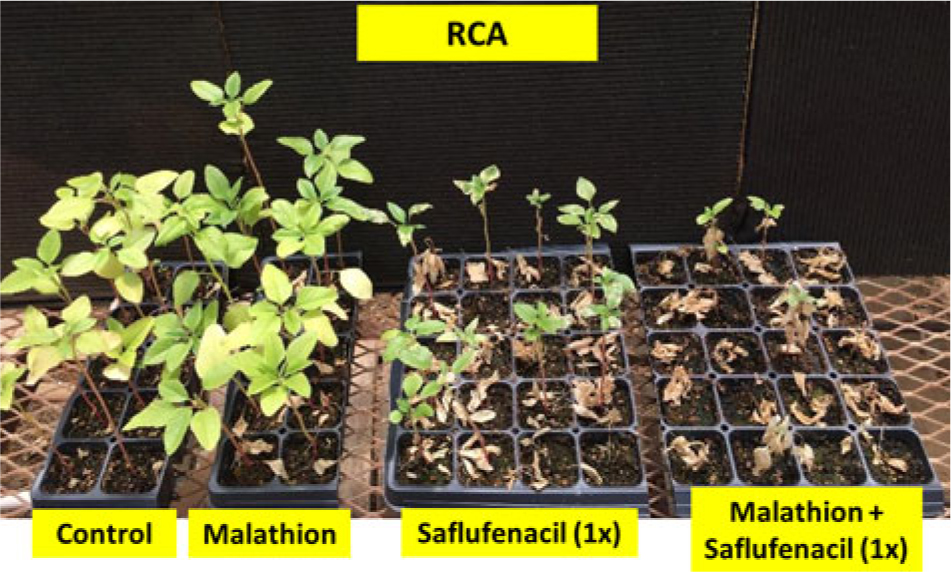
Figure 2. Palmer amaranth Randolph County, AR (RCA), accession treated with P450 inhibitor malathion (1,500 g ai ha−1), saflufenacil (410 g ai ha−1), or malathion followed by saflufenacil. Photograph taken 2 wk after treatment.
Resistance Profile of the RCA Accession
Nontarget site resistance confers cross-resistance to different herbicides within the same SOA group (Délye etal. Reference Délye, Jasieniuk and Le Corre2013; Vencill etal. Reference Vencill, Nichols, Webster, Soteres, Mallory-Smith, Burgos, Johnson and McClelland2012). Therefore, the cross- and multiple-resistance profiles of the RCA accession were tested against different herbicides applied at field rates and evaluated 2 WAT (Table 1). Treatment with pyrithiobac sodium showed the highest percent survival (94.7%) fb acifluorfen (78.7%), tembotrione (69.3%), flumioxazin (60%), and saflufenacil (34.7%) (Table 3). In comparison, Palmer amaranth survival was 0% and 4% for dicamba and atrazine applications, respectively, indicating the RCA accession is still sensitive to these herbicides (Table 3). The susceptible check (S2) used in this experiment was completely controlled by the aforementioned herbicides with 0% survival, when compared with a nontreated control (Table 3). In general, percent biomass (relative to nontreated plants) exhibited similar trends with percent survival. These findings indicate the fomesafen-resistant RCA accession is cross-resistant to other PPO-inhibiting herbicides (i.e., flumioxazin, acifluorfen, saflufenacil) and has multiple resistance to HPPD (i.e., tembotrione) and ALS (i.e., pyrithiobac sodium) inhibitors, and glyphosate. Because of the wide variability in Palmer amaranth accessions, resistance to the HPPD inhibitors mesotrione and tembotrione was found to be widespread in Arkansas and is most likely due to a nontarget site–based mechanism (Singh etal. Reference Singh, Burgos, Singh, Alcober, Salas-Perez and Shivrain2018).
Table 3. Cross-resistance profile of Palmer amaranth RCA accession to different herbicide modes of action.a

a Herbicides were applied to RCA seedlings at the four- to six-leaf stage and evaluated 2 wk after treatment. The greenhouse experiment was conducted at the Altheimer Laboratory, University of Arkansas, Fayetteville.
b Abbreviations: ALS, acetolactate synthase; HPPD, 4-hydroxyphenylpyruvate dioxygenase; PPO, protoporphyrinogen oxidase; PSII, photosystem II; RCA, Randolph County, AR; S2, 1986 susceptible check.
c Means with no common letter(s) within a column are significantly different according to Fisher’s protected LSD test (P < 0.05).
d Percent biomass relative to nontreated control.
e Application rate: 160 g ai ha−1.
f Application rate: 410 g ai ha−1.
g Application rate: 420 g ai ha−1.
h Application rate: 92 g ai ha−1.
i Application rate: 1,120 g ai ha−1.
j Application rate: 560 g ae ha−1.
k Application rate: 72 g ai ha−1.
ALS Gene Sequencing
The ALS gene was sequenced from RCA plants that survived the 1× (72 g ai ha−1) application rate of pyrithiobac sodium. No point mutations at positions 122, 197, 205, 376, 377, 574, 653, and 654 in the ALS enzyme were identified in the RCA accession. The mutations at eight locations spread over five different domains (A to E) in the ALS enzyme are known to confer varying levels of ALS-inhibitor resistance in weed species (Tranel and Wright Reference Tranel and Wright2002; Yu and Powles Reference Yu and Powles2014). It is concluded that a target-site mechanism based on a point mutation is not involved in conferring pyrithiobac resistance in the RCA accession, and resistance may be a consequence of increased cytochrome P450 activity selected by PPO inhibitors or by prior exposure to ALS inhibitors. Guo etal. (Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015) reported similar results with an A. tuberculatus population resistant to HPPD and PSII inhibitors.
Effect of Malathion on Control with PPO Herbicides
Varanasi etal. (Reference Varanasi, Brabham and Norsworthy2018b) had reported that malathion partially reversed fomesafen resistance in the RCA accession. The present study investigated the response of the RCA accession to malathion with and without other PPO-inhibiting herbicides. Treatment with malathion fb saflufenacil resulted in a decrease in the survival rate (23%; P < 0.05) compared with saflufenacil applied alone (63%; P < 0.05) (Table 4; Figure 2). On the contrary, no reduction in percent survival and biomass was observed for RCA accession after malathion fb flumioxazin or malathion fb acifluorfen treatments, compared with herbicides applied alone (Table 4). Similar to our earlier studies with fomesafen (Varanasi etal. Reference Varanasi, Brabham and Norsworthy2018b), the results show that malathion improved control with saflufenacil by partially reversing the PPO-resistant phenotype (Figure 2). Organophosphate insecticides such as malathion and synergists target different classes of cytochrome P450s; therefore, it is likely that a different set of cytochrome P450s are involved in conferring metabolic resistance to flumioxazin and acifluorfen compared with saflufenacil and fomesafen. Additional investigations using radiolabeled herbicides are necessary to confirm these results.

a Flumioxazin, acifluorfen, and saflufenacil.
b Herbicides were applied to RCA seedlings at the four- to six-leaf stage and evaluated 2 wk after treatment. The greenhouse experiment was conducted at the Altheimer Laboratory, University of Arkansas, Fayetteville, AR.
c Abbreviations: fb, followed by; PPO, protoporphyrinogen oxidase; RCA, Randolph County, AR.
d Means with no common letter(s) within a column are significantly different according to Fisher’s protected LSD test (P < 0.05).
e Percent biomass relative to nontreated control.
f Application rate: 1,500 g ai ha−1.
g Application rate: 160 g ai ha−1.
h Application rate: 420 g ai ha−1.
i Application rate: 410 g ai ha−1.
Soybean Response to Fomesafen, P450, and GST Inhibitors
Previous research has suggested the use of cytochrome P450 and GST inhibitors as alternative approaches to manage nontarget-site herbicide resistance in weed species (Cummins etal. Reference Cummins, Wortley, Sabbadin, He, Coxon, Straker, Sellars, Knight, Edwards, Hughes, Kaundun, Hutching, Steel and Edwards2013; Pan etal. Reference Pan, Si, Yu, Tu and Powles2012). Therefore, the effects of fomesafen with P450 and GST inhibitors on soybean injury were evaluated to verify whether these compounds could be used under field conditions to manage NTSR. No reduction relative to the nontreated plants was observed in soybean fresh biomass 2 WAT with malathion (98.6%) or NBD-Cl (96.1%) (Table 5). Similarly, no biomass reduction occurred when plants were treated with malathion plus fomesafen (89.2%), NBD-Cl plus fomesafen (85.9%), and fomesafen (81.3%) alone, showing that application of P450 or GST inhibitors in combination with fomesafen does not lead to significantly adverse effects on soybean (Table 5). The P450/GST inhibitor treatments in combination with fomesafen caused initial injury (10%) to the plants; however, the injury was transient, and plants recovered within a week.
Table 5. Effect of fomesafen, cytochrome P450 (malathion), and glutathione S-transferase (NBD-Cl) inhibitors on soybean.a

a Treatments were applied at V2 stage of soybean and evaluated 2 wk after treatment. The greenhouse experiment was conducted at the Altheimer Laboratory, University of Arkansas, Fayetteville, AR.
b Abbreviation: NBD-Cl, 4-chloro-7-nitrobenzofurazan.
c Percent biomass relative to nontreated control.
d Means with no common letter(s) within a column are significantly different according to Fisher’s protected LSD test (P < 0.05).
e Application rate: 1,500 g ai ha−1.
f Application rate: 270 g ai ha−1.
g Application rate: 395 g ai ha−1.
In conclusion, the nontarget-site fomesafen-resistant Palmer amaranth accession exhibited cross-resistance to selected PPO-, HPPD-, and ALS-inhibiting herbicides (i.e., flumioxazin, acifluorfen, saflufenacil; tembotrione; and pyrithiobac sodium, respectively) in addition to glyphosate. These data lead to further validation of earlier studies showing unpredictable resistance to different herbicide chemistries due to NTSR (Délye etal. Reference Délye, Gardin, Boucansaud, Chauvel and Petit2011). Similar to our previous research with fomesafen (Varanasi etal. Reference Varanasi, Brabham and Norsworthy2018b), the cytochrome P450 inhibitor malathion enhanced the effect of saflufenacil in the RCA accession. However, future metabolic research using radioisotopes (14C), analyses by gas chromatography–mass spectrometry, and liquid chromatography–mass spectrometry are needed to confirm these findings and identify parent herbicide and metabolites. Identification of the cytochrome P450 gene(s) conferring metabolic resistance to fomesafen and saflufenacil are key components toward understanding the mechanism and evolution of NTSR. Prevalence of cross- and multiple resistance in Palmer amaranth accessions emphasizes the need for adoption of integrated weed management strategies in combating the evolution of herbicide-resistant weeds. This work highlights the need for new research approaches toward the important, but still relatively unknown, nontarget-site herbicide resistance.
Author ORCIDs
Vijay K. Varanasi, https://orcid.org/0000-0002-3571-5263
Acknowledgement
Funding for this research was provided by the Arkansas Soybean Promotion Board. No conflicts of interest have been declared.









