Introduction
It is widely thought that virtually every free-living species is infected by at least one parasite species, highlighting the substantial contribution of parasites to biodiversity (Poulin and Morand, Reference Poulin and Morand2000). Numerous examples exist of a single host species hosting multiple parasite species, as well as instances where one parasite species either inhabits multiple host species or requires multiple host species to complete its life cycle; all of these possibilities make it challenging to tease apart host–parasite interactions (Rigaud et al., Reference Rigaud, Perrot-Minnot and Brown2010; Pilosof et al., Reference Pilosof, Morand, Krasnov and Nunn2015). When multiple parasite species coexist in one host individual, competition for resources is expected (Dobson, Reference Dobson1985; Mideo, Reference Mideo2009), because parasites by definition rely on their host for survival during at least part of their life cycle. Niche partitioning is an outcome of parasites avoiding competition when they contemporaneously co-occur within one host individual (Bashey, Reference Bashey2015). Detailed observations of locations within a host combined with comparative molecular data have revealed several different cases of niche partitioning among parasites occurring within the same host individual (Wakeman et al., Reference Wakeman, Heintzelman and Leander2014; Mohapatra et al., Reference Mohapatra, Sura and Seth2022).
Annelids play important ecological roles in marine environments as both predators and prey for other animals, and also contribute to sediment bioturbation (Hutchings, Reference Hutchings1998). Furthermore, annelids host a diverse range of animal and protistan symbionts (Peoples et al., Reference Peoples, Randhawa and Poulin2012), where gregarine apicomplexans are particularly common within the host intestines (Leander, Reference Leander2008; Schrével et al., Reference Schrével, Valigurová, Prensier, Chambouvet, Florent and Guillou2016; Rueckert and Horák, Reference Rueckert and Horák2017; Paskerova et al., Reference Paskerova, Miroliubova, Diakin, Kováčiková, Valigurová, Guillou, Aleoshin and Simdyanov2018; Iritani et al., Reference Iritani, Banks, Webb, Fidler, Horiguchi and Wakeman2021). Gregarines are commonly known as parasites; however, they might show a broader spectrum of symbioses ranging from mutualism to parasitism (Rueckert et al., Reference Rueckert, Betts and Tsaousis2019). Gregarines are known for their host specificity and are typically horizontally transmitted through the faecal-oral route (Leander, Reference Leander2008; Lynn, Reference Lynn2008; Desportes and Schrével, Reference Desportes and Schrével2013). The infestation process in a new host animal begins when the host ingests oocysts (syn., sporocysts) from the environment. Then, four or more haploid sporozoites (the infectious stage) emerge from an oocyst and penetrate the gut epithelial cells of the host. The sporozoites develop into larger feeding cells, called ‘trophozoites’, that emerge from host cells and occupy extracellular spaces within the host, such as the intestinal lumen. Two mature trophozoites subsequently pair up in a process known as ‘syzygy’ (the mating stage) to form a connected pair of gamonts that are eventually enveloped by a cyst wall, forming a ‘gametocyst’. The two gamonts within the gametocyst undergo a series of cell divisions to form female and male gametes that fuse together to produce a fleeting diploid sporoblast (i.e. zygote). Each sporoblast develops into an oocyst, within which meiosis occurs to from four (or more) resting sporozoites. Therefore, each gametocyst can be filled with hundreds of oocysts that ultimately leave the host via the faeces and become distributed in the surrounding sediments. Trophozoites are the most conspicuous, dynamic and morphologically diverse stage in the life cycle of gregarines, so they commonly form the basis for species discovery and identification (Leander, Reference Leander2008).
Since the early stages of gregarine research, it has been well-known that multiple infections of gregarines and other protists can occur within an individual annelid host (Ray, Reference Ray1930; Fowell, Reference Fowell1936). In the modern era, small subunit (SSU) rDNA sequencing has become a widely used method for species identification and confirming the presence of multiple parasite species within a host (Leander et al., Reference Leander, Clopton and Keeling2003; Wakeman et al., Reference Wakeman, Heintzelman and Leander2014; Diakin et al., Reference Diakin, Paskerova, Simdyanov, Aleoshin and Valigurová2016; Rueckert and Horák, Reference Rueckert and Horák2017; Iritani et al., Reference Iritani, Banks, Webb, Fidler, Horiguchi and Wakeman2021; Paskerova et al., Reference Paskerova, Miroliubova, Valigurová, Janouskovec, Kováciková, Diakin, Sokolova, Mikhailov, Aleoshin and Simdyanov2021). Some detailed studies have demonstrated that different parasites can occupy distinct locations within the host's body or different life stages of the host. For instance, in the yellow mealworm Tenebrio molitor, three gregarine species were exclusively detected in the larval stage, while one species exclusively infected the adult stage, indicating temporal niche partitioning (Clopton et al., Reference Clopton, Janovy and Percival1992). In the case of the common heart urchin Echinocardium cordatum, five gregarine species were discovered, with three inhabiting the intracoelomic space and 2 found within the intrahemal space (Coulon and Jangoux, Reference Coulon and Jangoux1986). In the spaghetti worm Thelepus japonicus, Selenidium terebelle and S. melongena occupied the intestinal and coelomic spaces, respectively, demonstrating another case of spatial niche partitioning (Wakeman et al., Reference Wakeman, Heintzelman and Leander2014).
In addition to gregarines, other parasitic alveolates (e.g. dinoflagellates, ciliates, protococcidians, rhytidocystids and squirmids) have also been discovered in marine annelids (Kozloff, Reference Kozloff1961; Leander, Reference Leander2006; Leander and Ramey, Reference Leander and Ramey2006; Park et al., Reference Park, Cooney, Phua, Horiguchi, Husnik, Keeling, Wakeman and Leander2023; Paskerova et al., Reference Paskerova, Miroliubova, Valigurová, Aleoshin and Simdyanov2023). Among ciliates, several species of astomes have been documented in marine annelid hosts (Caullery and Mesnil, Reference Caullery and Mesnil1907; Awerinzew, Reference Awerinzew1908; De Puytorac, Reference De Puytorac1954; Sauvadet et al., Reference Sauvadet, Lynn, Roussel, Le Panse, Bigeard, Schrével and Guillou2017). As their name suggests, astomes lack a cytostome (i.e. a cell mouth) (De Puytorac, Reference De Puytorac1954; Lynn, Reference Lynn2008) and are thought to be commensal inhabitants within the intestines of diverse animals, including molluscs, turbellarians, newts and frogs (Fokam et al., Reference Fokam, Ngassam, Strüder-Kypke and Lynn2011; Obert and Vďačný, Reference Obert and Vďačný2019). While more than 50 genera from 9 families of astomes are known, molecular phylogenetic data remain limited and are only available for astomes that inhabit oligochaetes (Fokam et al., Reference Fokam, Ngassam, Strüder-Kypke and Lynn2011; Obert and Vďačný, Reference Obert and Vďačný2019, Reference Obert and Vďačný2021), polychaetes (Sauvadet et al., Reference Sauvadet, Lynn, Roussel, Le Panse, Bigeard, Schrével and Guillou2017), turbellarians (Rataj and Vďačný, Reference Rataj and Vďačný2018; Rataj et al., Reference Rataj, Zhang and Vd'ačný2022) and gastropods (Zhang and Vd'ačný, Reference Zhang and Vd'ačný2022). Among marine species, SSU rDNA sequences are currently accessible for only a few species of Durchoniella (astome ciliate) that infect cirratulid polychaete Cirriformia (Sauvadet et al., Reference Sauvadet, Lynn, Roussel, Le Panse, Bigeard, Schrével and Guillou2017).
Myxicola Koch in Renier 1847 (Polychaeta: Sabellidae), which comprises more than 20 named species of feather duster worms, is a group of marine annelids known for producing gelatinous substances and exhibiting remarkable regenerative abilities (Stabili et al., Reference Stabili, Schirosi, Licciano and Giangrande2014; Licciano et al., Reference Licciano, Watson, Murray and Giangrande2015). While exploring various symbionts in marine annelid hosts collected from Harriot Bay, Quadra Island (British Columbia, Canada), we identified two different Selenidium species (Apicomplexa, Gregarinida) and one astome ciliate species in two species of Myxicola: Myxicola. sp. Quadra (resembling M. infundibulum) and M. aesthetica. We utilized SSU rDNA sequences, light microscopy (LM), histology and scanning electron microscopy (SEM) to characterise the endosymbionts found in these Myxicola hosts. We compared the traits of the Selenidium species described in this study with the traits of Selenidium mesnli Brasil, Reference Brasil1909, which was described over 100 years ago from individuals of Myxicola infundibulum collected in Europe (Brasil, Reference Brasil1909; Ray, Reference Ray1930; Reed, Reference Reed1933). Based on comparative morphology and molecular phylogenetic data from the symbionts observed in this study, we identify Selenidium cf. mesnili and establish one new species of gregarine apicomplexans and one new species of astome ciliates from the Pacific Ocean: Selenidium elongatum n. sp. and Pennarella elegantia n. gen. et sp.
Materials and methods
Sample collection and light microscopy
Various numbers (10–40) of specimens of Myxicola (Fig. 1) were collected by SCUBA diving in November 2021; January, February, April, October and November 2022; and in March and August 2023. Two different species of Myxicola were present in these collections: ‘Myxicola sp. Quadra’ (resembling M. infundibulum) and M. aesthetica. Specimens of Myxicola sp. Quadra were discovered on the sediment near Cove Beach (50°07′00.8″N 125°13′20.9″W), while specimens of M. aesthetica were found on ropes connected to a raft located ~10–20 m away from the collection site of Myxicola sp. Quadra (50°07′01.5″N 125°13′22.0″W). The two species of Myxicola were clearly distinguished by their size. Myxicola sp. Quadra measured 5–10 cm in length and only a single individual was found within a gelatinous incasement. In contrast, M. aesthetica specimens measured 1.5–3 cm in length, and most individuals (n = 10–30) were discovered within a mass of gelatinous material (Fig. 1A–D).
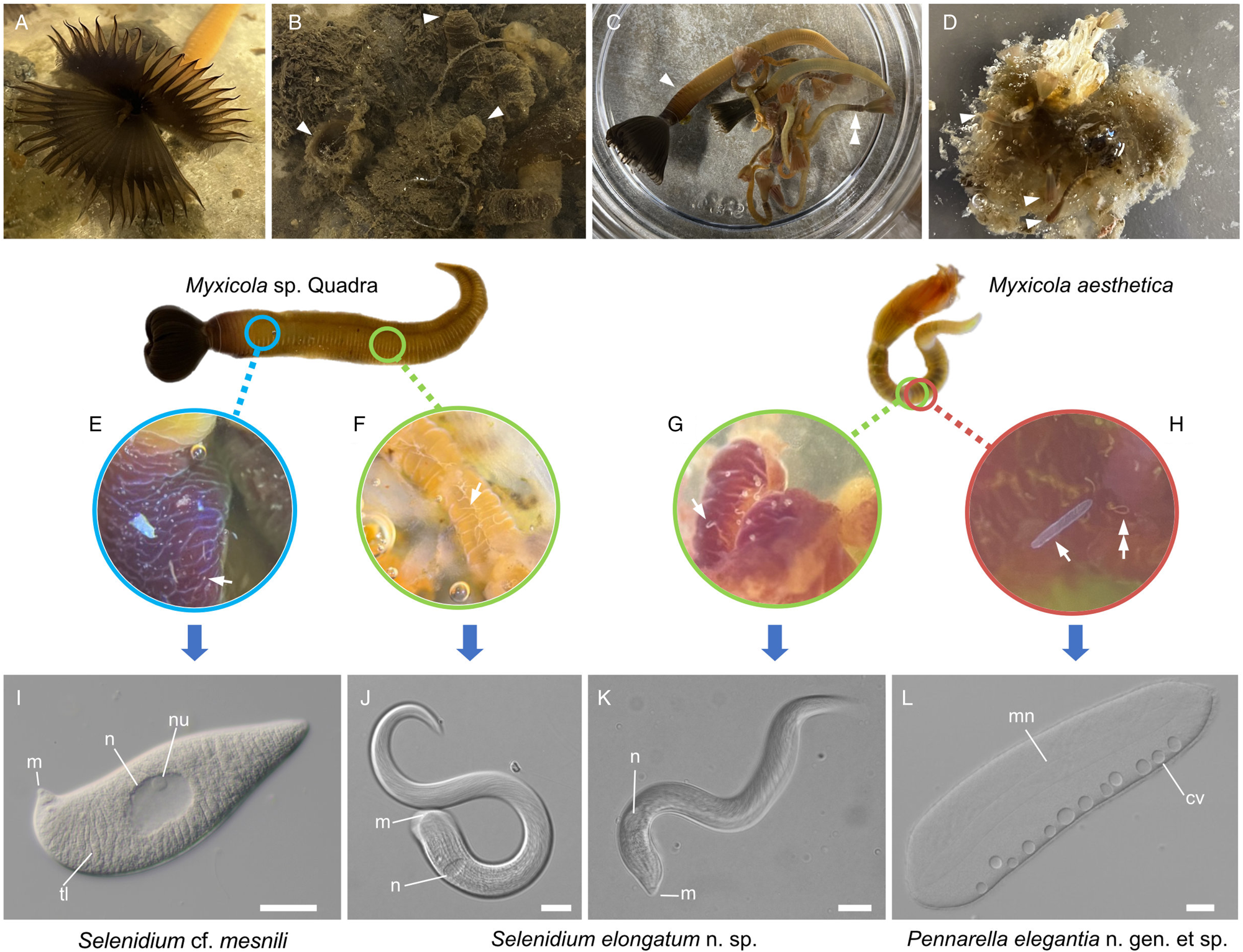
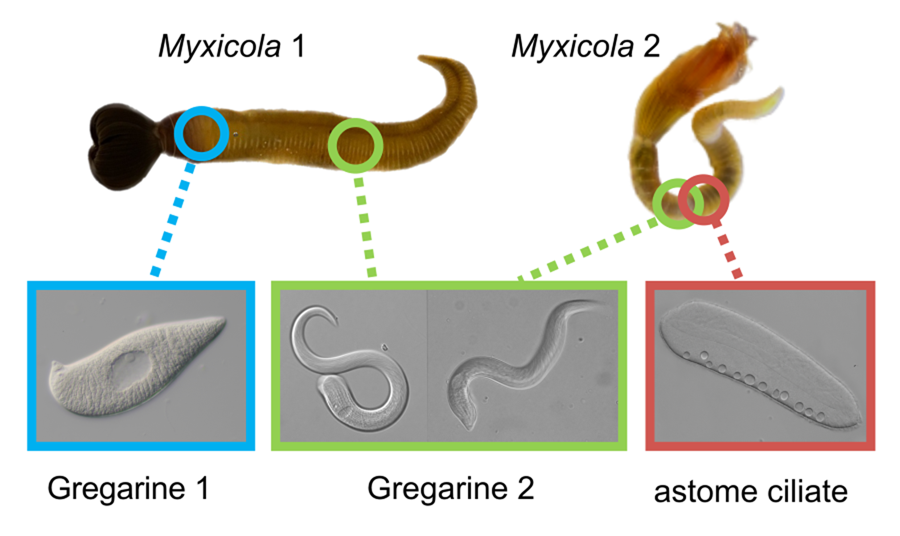
Figure 1. Two gregarine species and one ciliate species found in two Myxicola host species. (A) A live specimen of Myxicola sp. Quadra placed in a container filled with seawater. The brown-coloured crown, used for respiration and food collection, is visible. (B) Specimens of Myxicola sp. Quadra in gelatinous substance produced by them. A few individuals are indicated with arrowhead. (C) The two Myxicola species investigated in this study. Myxicola sp. Quadra (single arrowhead) is larger than M. aesthetica (double arrow head). (D) A colony of M. aesthetica in the gelatinous substance produced by them. A few individuals are indicated with arrowhead. (E) A large number of small, lemon-shaped trophozoites of Selenidium cf. mesnili (arrow) were visible on the epithelium of the foregut of Myxicola sp. Quadra. (F) Several long trophozoites of Selenidium elongatum n. sp. (arrow) were found in the mid to hindgut of Myxicola sp. Quadra. (G) Several Selenidium elongatum n. sp. (arrow) were attached to the epithelium of the gut of M. aesthetica. (H) Free swimming Pennarella elegantia n. gen. et sp. (arrow) inhabited the gut of M. aesthetica. Selenidium elongatum n. sp. coinfecting the same host are visible in the background (double arrowhead). I. Differential interference contrast (DIC) light micrograph of Selenidium cf. mesnili (scale bar = 5 μm). (J) DIC light micrograph of Selenidium elongatum n. sp. from Myxicola sp. Quadra (scale bar = 20 μm). (K) DIC light micrograph of Selenidium elongatum n. sp. from M. aesthetica (scale bar = 20 μm). (L) DIC light micrograph of astome ciliate Pennarella elegantia n. gen. et sp. from M. aesthetica (scale bar = 20 μm). n, nucleus; nu, nucleolous; m, mucron; tl, transverse lines; mn, macronucleus; cv, contractile vacuoles.
The collected animals were either dissected at a marine station (e.g., the Hakai Institute) near the collection sites or transported to a laboratory at the University of British Columbia (UBC), Canada. The animals were dissected using fine-tipped forceps and a scalpel, after which the intestine was carefully placed into a clean Petri dish containing filtered seawater. The intestine was opened, starting from the anterior end towards the posterior end, using fine-tipped forceps and a scalpel. Individual symbiont cells for PCR analysis were isolated using a hand-drawn glass pipette and subsequently washed three times in filtered seawater. Endosymbionts with different morphotypes were then separated and transferred to a glass slide for examination under differential interference contrast (DIC) microscopy. A Sony ILC-7RM4A digital camera attached to Zeiss Axio Scope A1 compound microscope and Zeiss Axiocam 503 colour camera attached to Zeiss Axioplan 2 microscope or Axiovert 200 inverted scope were used for micrographs at the marine station and at UBC, respectively.
Scanning electron microscopy and histology
Individual cells of Selenidium spp. and astome ciliates were picked up using a hand-drawn glass pipette and subsequently transferred to a Swinnex filter holder fitted with an isopore membrane (pore size = 10 μm), which was filled with filtered seawater. The cells were fixed with osmium vapour for 10 min, followed by the addition of 6–7 drops of 4% osmium tetroxide directly into the Swinnex filter holder for post-fixation for 10 min. The osmium-fixed cells were rinsed twice with filtered seawater. Ethanol dehydration was performed in a series, starting with 30–50%, 70%, 80%, 85%, 90%, 95%, and finally 100% ethanol. The membrane was transferred to a metal basket submerged in 100% ethanol for critical point drying using the Tousimis autosamdri 815B Critical Point Dryer. Once dried, the membrane with specimens was placed onto a stub and a 2 nm layer of platinum gold was sputter coated using either the Cressington 208HR high-resolution sputter coater or the Leica EM ACE600 Coater. Images were captured using Zeiss Crossbeam XB350. SEM processing and imaging were conducted at the UBC Bioimaging Facility.
For histological observations, a whole intestine was isolated from a specimen of Myxicola sp. Quadra. The anterior and mid-to-hind gut parts were then separately placed into 1.5 mL tubes. The tissues were fixed in 4% paraformaldehyde (PFA) overnight and stored in 70% ethanol at 4°C until they were sent to the Diagnostic & Research Histology Laboratory at UBC for histological analysis. The tissues were embedded in paraffin blocks, and 4 μm-thick sections were stained with haematoxylin and eosin (H&E). Imaging was performed using a Zeiss Axiocam 503 colour camera attached to a Zeiss Axioplan 2 microscope.
PCR and sequencing
Trophozoites representing different morphotypes of Selenidium were individually isolated from the host, washed 3 times in filtered seawater and placed into an Eppendorf tube. 10 μL of distilled water was added to each tube containing a single trophozoite of Selenidium. The tubes were then incubated at 56°C for 3 min to obtain template DNA for PCR. To obtain SSU rDNA sequences of Selenidium, two consecutive PCR runs were conducted for nested PCR. For the first round of PCR, 22 μL of distilled water, 0.5 μL each of forward and reverse primers (GF2; 5′-TGCGCTACCTGGTTGATCC-3′ and SSUR4; 5′-GATCCTTCTGCAGGTTCACCTAC-3′) and 2 μL of DNA template were added in a 0.2 mL tube containing a PCR bead (Cytiva PuReTaq Ready-To-Go™ PCR Beads; Cytiva, Global Life Sciences Solutions, Marlborough, MA, USA). PCR reactions were conducted with the following conditions: an initial denaturation at 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 90s and a final extension at 72°C for 7 min. The second round of PCR was conducted using 23 μL of distilled water, 0.5 μL each of forward and reverse primers (GF5; 5′-CCTGGTTGATCCTGCCAG-3′ and SEL_R1; 5′-GAAACCTTGTTACGACTTCTCC-3′), 1 μL of PCR product from the first round of PCR, and a PCR bead with the following conditions: an initial denaturation at 95°C for 3 min, 26 cycles of 95°C for 30s, 52°C for 30s, 72°C for 90 s and a final extension at 72°C for 7 min. SSU rDNA sequences of astome ciliates were obtained using the primer pair GF5 and SSUR4 with the following conditions: an initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 90 s and a final extension for 7 min at 72°C.
To obtain COI sequences of the host specimens, 12.3 μL of distilled water, 4 μL of reaction buffer, 0.8 μL of each forward and reverse primer (LCO1490 and HCO2198; Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994), 0.1 μL of MyTaq (Bioline) and 2 μL of DNA were used. PCR conditions were as follows: an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 48°C for 1 min, 72°C for 40 s and a final extension for 10 min at 72°C. Afterward, 5 μL of PCR product from each reaction was loaded onto a 1.5% agarose gel stained with GelRedTM (Biotium) and subsequently purified using ExoSAP-ITTM (Applied Biosystems). The purified PCR products were sent to the UBC sequencing and bioinformatics facility for sequencing.
Molecular phylogenetic analyses
Three separate DNA sequence datasets were created for Selenidium, astome ciliates and Myxicola hosts, respectively. Selenidium is not a monophyletic group when inferred from SSU rDNA sequences but consists of several genetically divergent lineages (Rueckert and Horák, Reference Rueckert and Horák2017). The Selenidium sequences obtained in this study were highly similar to those of the main Selenidium clade based on a BLAST search. The sequences of Selenidium showed identities ranging from 93.75% to 98.64% with ‘S. cf. mesnili’ (JN857968) and 92.44% to 93.90% with S. sabellae (MF882906). They demonstrated a 90.55–91.94% similarity with S. pendula (MF882901), the type species of the genus Selenidium. Therefore, for the Selenidium dataset, one SSU rDNA sequence representing each Selenidium species of the ‘main’ clade (i.e. the clade containing the type species of the genus) available in GenBank was included, and two blastogregarine sequences were used as outgroups. As a result, 25 SSU rDNA sequences were included in the dataset. For astome ciliates, one SSU rDNA sequence representing each astome ciliate species available in GenBank, as well as sequences representing major groups of Oligohymenophorea were included. As a result, 53 SSU rDNA sequences were included including three sequences for outgroups. For the Myxicola hosts, all COI sequences of the genus Myxicola available in GenBank (21 sequences), as well two sequences for outgroups, were included. In all three datasets, DNA sequences were aligned using the MAFFT algorithm (Katoh and Standley, Reference Katoh and Standley2013) within Geneious Prime. Ambiguous sites were subsequently removed for the SSU rDNA alignments using Gblocks (Castresana, Reference Castresana2000). As a result, alignments of 1574 bp, 1595 bp and 685 bp were used for further analyses for Selenidium, astome ciliates and Myxicola, respectively.
All phylogenetic analyses were conducted using CIPRES Science Gateway v3.3 (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). Maximum likelihood (ML) trees were inferred using RAxML v8.2.12 with GTRCAT approximation (Stamatakis, Reference Stamatakis2014). A rapid bootstrap analysis was conducted with 1000 replicates. The best-fitting model of nucleotide evolution for each dataset was determined based on the corrected Akaike information criterion (AICc) using jModelTest v2.1.6 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). As a result, GTR + I + G was chosen as the best model for Selenidium and astome datasets, and HKY + I + G was chosen for Myxicola. For Bayesian tree inference, two independent runs, consisting of four chains each, were simultaneously conducted for 1 000 000 generations with a sampling frequency of 1000. A stop rule was applied to terminate the MCMC generations as soon as the standard deviation of split frequencies fell below 0.01. The initial 25% of samples were discarded. The resulting trees were visualized using FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Coinfection and distribution of endosymbionts in the host
At least 20 individuals of Myxicola sp. Quadra and 30 individuals of M. aesthetica were observed. Nearly all (>90%) individuals of Myxicola sp. Quadra examined in this study hosted two different morphotypes of Selenidium trophozoites: a short morphotype (= S. cf. mesnili) and a long morphotype (= S. elongatum n. sp.). Within Myxicola sp. Quadra, the foregut region was heavily infested with the short morphotype of Selenidium, which appeared as white dots under a stereoscope (Fig. 1E). These short morphotype cells were either embedded in or attached to the gut epithelium. In contrast, only the long morphotypes of Selenidium, which resembled nematodes under a stereoscope, were observed in the mid to hindgut region of the host, where they were attached to the gut epithelium (Fig. 1F). Histological sections (Figs. 2B and 2C) confirmed the presence of the two morphotypes and their locations in the epithelial cells in the anterior part of the gut (for the short morphotype) and in the gut lumen of the mid-posterior part (for the long morphotype) in the host, consistent with the observations made using light microscopy. In M. aesthetica, one morphotype of Selenidium ( = S. elongatum n. sp.) trophozoites (Fig. 1G) and one astome ciliate species (= Pennarella elegantia n. gen. et sp.) (Fig. 1H) almost always occurred together within the gut of a single host individual. No specific distribution patterns of Selenidium trophozoites and the astome ciliates along the gut were observed; however, the Selenidium trophozoites were attached to the gut epithelium, while the astome ciliates were freely swimming within the gut lumen (Fig. 1G and H).

Figure 2. Comparison of trophozoites and syzygy of Selenidium from Myxicola using light micrographs (LMs) and previously reported drawings. (A) A drawing of trophozoites of Selenidium mesnili inside the epithelial cells of host specimens of Myxicola infundibulum collected in the UK (modified from Ray, Reference Ray1930). (B) A histological section of the anterior part of the gut of Myxicola sp. Quadra (Canada) showing trophozoites of Selenidium cf. mesnili inside host epithelial cells (Scale bar = 20 μm). (C) A histological section of the mid-part of the gut of Myxicola sp. Quadra (Canada) showing trophozoites of Selenidium elongatum n. sp (black arrowheads) in the gut lumen (Scale bar = 20 μm). (D) A drawing of S. mesnili gametocytes in syzygy from M. infundibulum hosts collected in the UK (modified from Reed, Reference Reed1933). The gamonts are connected (arrow) by their posterior ends (Scale bar = 20 μm). (E) A histological section showing gametocytes of S. cf. mesnili from Myxicola sp. Quadra (Canada) in syzygy within a host epithelial cell. The syzygy junction is indicated by the arrow (Scale bar = 10 μm). F. Native observations under DIC of the gamonts of S. cf. mesnili from Myxicola sp. (Quadra) in syzygy and connected (arrow) by their posterior ends (Scale bar = 10 μm). (G) Native observations under DIC showing syzygy of Selenidium elongatum n. sp. from Myxycola sp. Quadra (Scale bar = 20 μm). The syzygy junction is indicated by the arrow. B, C and E were obtained from a single host individual, while F and G were obtained from another single host individual. n, nucleus; nu, nucleolus; hn, nucleus of the host; gl, gut lumen; m, mucron.
Two different morphotypes of selenidium trophozoites
The most notable differences between the two morphotypes of Selenidium in Myxicola sp. Quadra were the cell lengths, the location of the nucleus and the presence or absence of cytoplasmic transverse lines (Fig. 1I and J, and Supplementary Fig. 1A and B). The short morphotype of Selenidium ( = S. cf. mesnili) in Myxicola sp. Quadra was 35 μm (26–45 μm, n = 13) long and 13 μm (8–23 μm, n = 13) wide. The transverse lines only seen in the short morphotype in Myxicola sp. Quadra were due to granules in the cytoplasm and were therefore only visible in the light micrographs (Fig. 1I). Although some intermediate-sized cells were observed, the position of the nucleus in the two morphotypes was distinct, with the short morphotype having the nucleus in the middle part of the trophozoite, while the long morphotype (= S. elongatum n. sp.) having the nucleus near the anterior end of the trophozoite. The nucleus of the short morphotype was ellipsoidal and longitudinally elongated, measuring 8.4 μm long (6–10 μm, n = 5) and 4.3 μm wide (4–5 μm, n = 5). The long morphotypes of Selenidium observed in both Myxicola sp. Quadra (Fig. 1J) and M. aesthetica (Fig. 1K and Supplementary Fig. 1C) were characterized by a relatively large cell size: 144 μm (98–200 μm, n = 6) long and 13 μm (8–23 μm, n = 7) wide in Myxicola sp. Quadra and 140 μm (100–185 μm, n = 6) long and 25 μm (20–30 μm, n = 6) wide in M. aesthetica. Additionally, the nucleus of the long morphotype was ellipsoidal and elongated transversely, positioned between the anterior end and the middle part of the trophozoite in both host species (Fig. 1J and 1K), with 6 μm long (5–10 μm, n = 8) and 12 μm wide (7–18 μm, n = 6) wide in Myxicola sp. Quadra, and 12 μm long (8–18 μm, n = 5) and 18.4 μm wide (18–20 μm, n = 5) in M. aesthetica. These trophozoites moved by twisting and bending. Moreover, the morphological traits of the long morphotypes of Selenidium in both hosts were indistinguishable when examined with SEM (Fig. 3A and 3B). The long morphotypes from both hosts displayed rounded or pointed mucrons (depending on their movement) along with 20–28 longitudinal epicytic folds and numerous epicytic transverse striations near the anterior or middle part of the cell (Fig. 3D and 3E). Because epicytic transverse striations were also visible on the short morphotype of Selenidium with SEM. (Fig. 3C), the most notable distinction between the two morphotypes under the SEM was relative cell length.
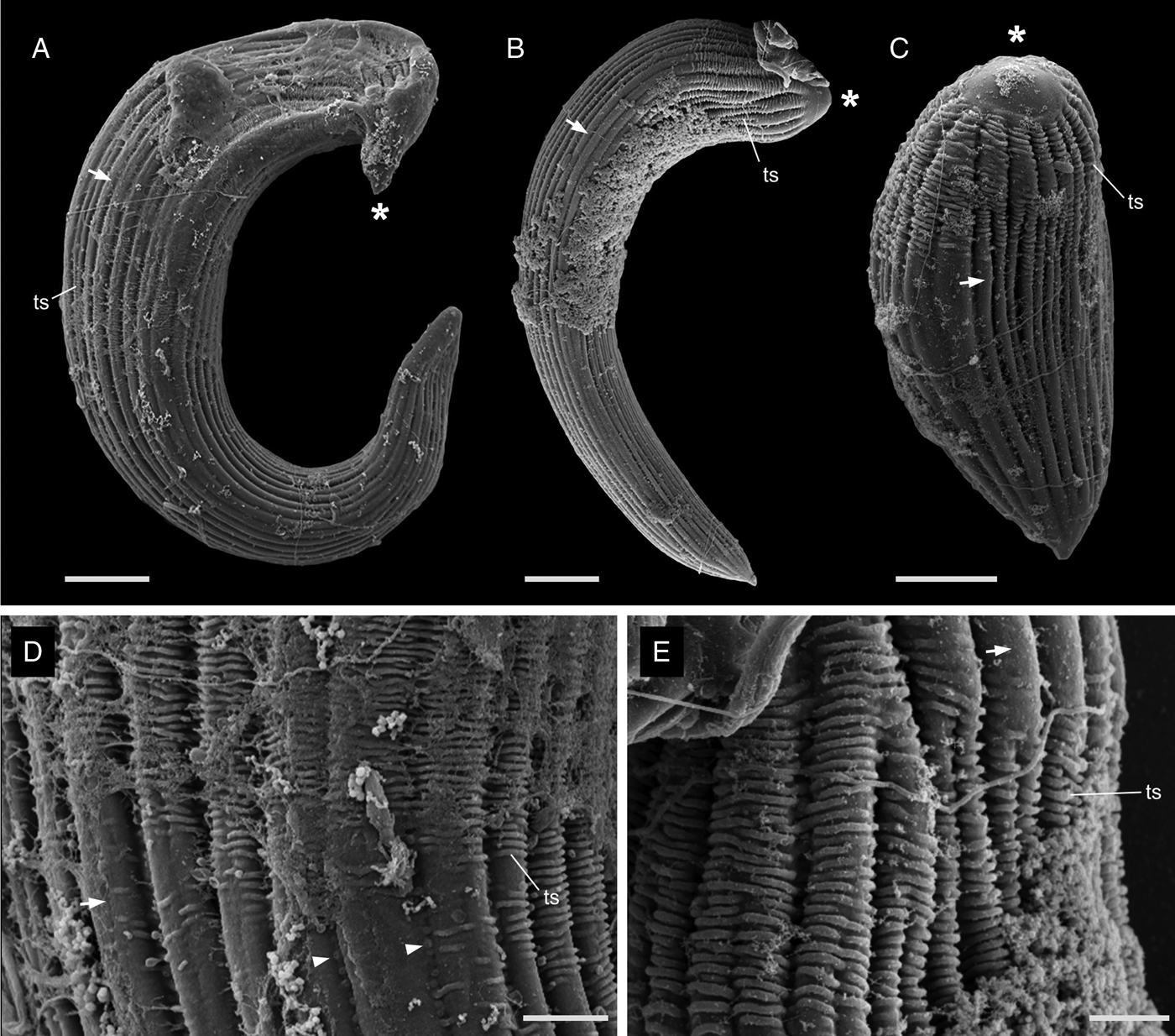
Figure 3. Scanning electron micrographs (SEM) of the trophozoites of Selenidium elongatum n. sp. and Selenidium cf. mesnili. (A) A trophozoite of Selenidium elongatum n. sp. obtained from Myxicola aesthetica showing a pointed mucron (*), longitudinal epicytic folds (arrow) and transverse striations (ts) (scale bar = 10 μm). (B) A trophozoite of Selenidium elongatum n. sp. from Myxicola sp. Quadra showing a rounded mucron (*), longitudinal epicytic folds (arrow) and transverse striations (ts) (scale bar = 10 μm). (C) A trophozoite of Selenidium cf. mesnili from Myxicola sp. Quadra showing the mucron (*), longitudinal epicytic folds (arrow) and transverse striations (ts) (scale bar = 5 μm). (D) High magnification SEM of a trophozoite of Selenidium elongatum n. sp. from M. aesthetica showing longitudinal epicytic folds (arrow), transverse striations (ts) and micropores within the epicytic grooves (arrowhead) (scale bar = 3 μm). (E) High magnification SEM of Selenidium elongatum n. sp. from Myxicola sp. Quadra showing transverse striations (ts) at the anterior end of the trophozoites. (scale bar = 2 μm).
Astome ciliates
Ciliates were observed swimming freely within the gut lumen of the host (Fig. 1H). The elongated cells were 267 μm (217–387 μm, n = 6) long and 70 μm (50–86 μm, n = 6) wide, with their surface densely covered by a multitude of cilia (Fig. 4). A macronucleus of 207 μm (150–320 μm, n = 4) long and 21 μm (20–25 μm, n = 4) wide was clearly visible, but a micronucleus was not detected in the light micrographs (Fig. 4A and Supplementary Fig. 1D). Typically, between 6 and 12 contractile vacuoles were arranged in a single row (Fig. 4A); however, sometimes, a second row of only 1–3 contractile vacuoles was visible on the opposite side of the cell. No attachment apparatus, such as a hook or an apparent thigmotactic area, was observed (Fig. 4A–C, and Supplementary Fig. 2).

Figure 4. Light and scanning electron micrographs (SEM) of the astome ciliate Pennarella elegantia n. gen. et sp. (A) A differential interference contrast (DIC) light micrograph showing a long, central macronucleus (mn) and a single row of contractile vacuoles (cv) (scale bar = 20 μm). (B) Low magnification SEM showing a single astome cell (scale bar = 20 μm). (C) High magnification SEM of the same cell showing the uniform array of cilia (scale bar = 4 μm).
Molecular phylogeny
Selenidium
Both ML and Bayesian analyses were conducted on the alignment of 1,574 bp from 25 taxa. The results showed that all of the Selenidium sequences obtained in this study belong to the largest Selenidium clade that includes the type species, S. pendulum (Fig. 5). All the Selenidium sequences obtained in this study, along with the sequence reported as Selenidium cf. mesnili (GenBank ID: JN857968) from Myxicola sp. collected in Victoria (Canada), formed a monophyletic group with strong support (BS = 100 and PP = 1). The long morphotype Selenidium from Myxicola sp. Quadra, the Selenidium isolated from M. aesthetica and the Selenidium isolated from an unverified species of Myxicola (reported as ‘Myxicola infundibulum’ in Wakeman and Leander, Reference Wakeman and Leander2012) exhibited high pairwise genetic similarities ranging between 97.8% and 99% (Table 1). However, the short morphotype of Selenidium from Myxicola sp. Quadra displayed less than 95% genetic divergence from the long morphotype of Selenidium (Table 1).
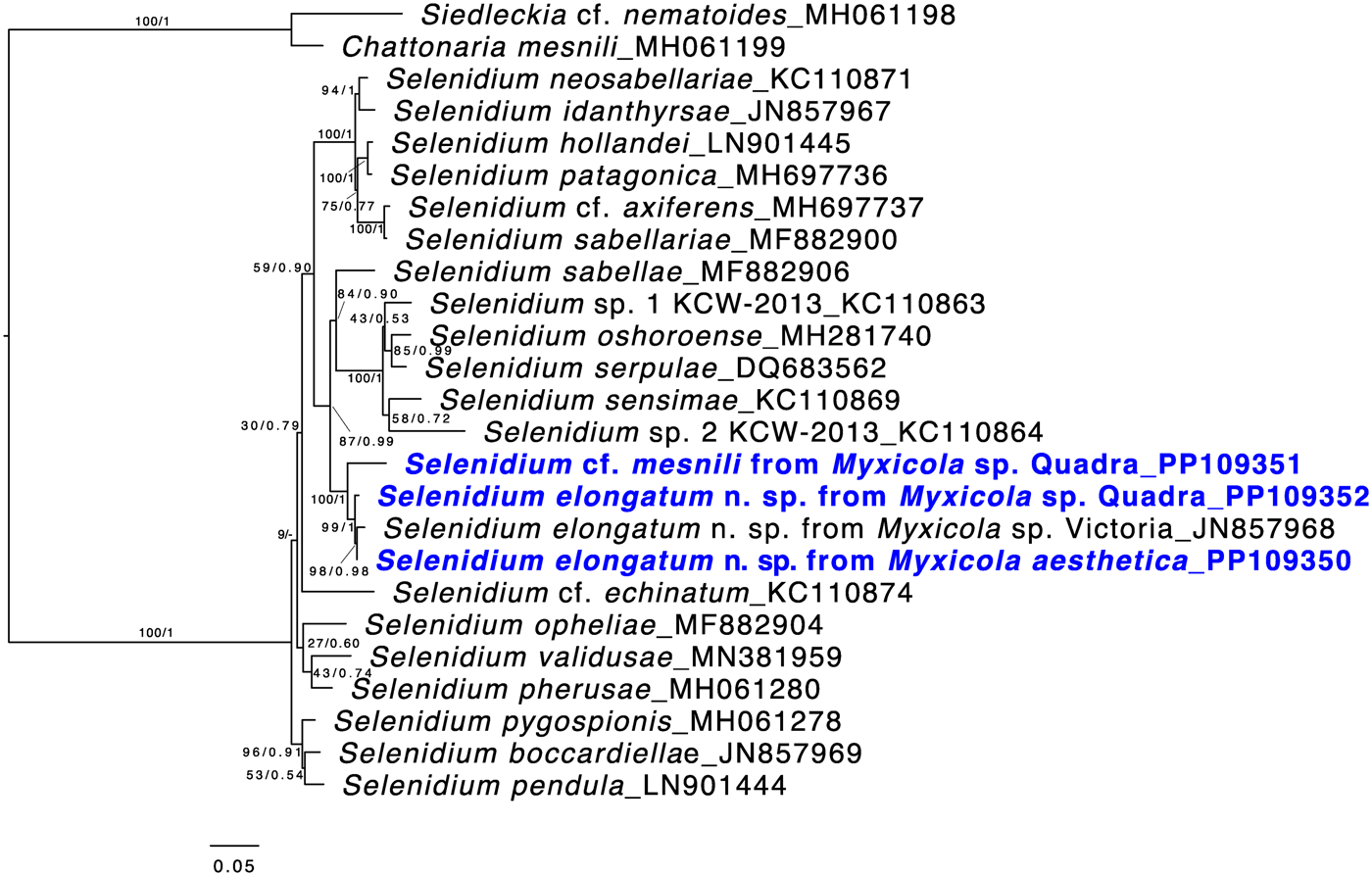
Figure 5. Maximum likelihood (ML) tree of the main lineage of Selenidium that includes the type species S. pendula. This tree was inferred from 25 small subunit (SSU) rDNA sequences and 1,574 unambiguously aligned sites using RAxML. Posterior probabilities (PP) and ML bootstrap supports (BS) are shown. Sequences from Selenidium obtained in this study are highlighted in bold blue font. All Selenidium species from Myxicola hosts form a monophyletic group with maximal support. Additionally, the clade containing all SSU sequences of Selenidium elongatum n. sp. (the long morphotype) is strongly supported, while the SSU sequence from Selenidium cf. mesnili (the short morphotype) is distinct from the others.
Table 1. Pairwise genetic distance of the small subunit rDNA sequences obtained from the Selenidium species inhabiting Myxicola host species

a Reported as S. cf. mesnili in Wakeman and Leander (Reference Wakeman and Leander2012).
Astome ciliates
Our ML and Bayesian analyses of the alignment of 1595 bp from 53 taxa strongly supports the major subgroups within Oligohymenophorea, except for Scuticociliatia (Fig. 6). Astomes from oligochaetes (earthworms) and marine polychaetes formed a monophyletic group with strong support (BS = 94 and PP = 1). However, Haptophrya planariarum, a ciliate found in flatworm hosts, did not cluster with astomes from annelid hosts but instead was grouped with scuticociliatians. Based on comparisons of SSU rDNA sequences, the astome ciliate species from M. aesthetica was 93.7% identical to Durchoniella species. The astomes from Myxicola appears to be sister to the clade containing all other annelid-infecting astomes; however, their relationships were poorly resolved (Fig. 6).

Figure 6. Maximum likelihood (ML) tree of oligohymenophorean ciliates showing the phylogenetic position of the new astome Pennarella elegantia n. gen. et sp. This tree was inferred from 53 small subunit (SSU) rDNA sequences and 1,595 unambiguously aligned sites using RaxML. Posterior probabilities (PP) and ML bootstrap supports (BS) are shown. The SSU rDNA sequence of the astome ciliate obtained in this study is highlighted in bold blue font. Major groups of Oligohymenophrea are marked with grey bars. Family names (blue bars) and host group information (pink bars) of astomes are shown. Although Astomatida is not a monophyletic group, the clade containing all of the annelid-infecting species is strongly supported, with polychaete-infecting species (i.e. Pennarella elegantia n. gen. et sp. and Durchoniella spp.) branching before the well supported oligochaete-infecting astome ciliates.
Myxicola hosts
Both ML and Bayesian analyses of the alignment of 685 bp of 23 taxa showed similar topologies (Fig. 7). Myxicola sp. Quadra showed 100% identity with two COI sequences obtained from Bamfield, Canada (HM473471-2; Carr et al., Reference Carr, Hardy, Brown, MacDonald and Hebert2011). All six COI sequences of M. aesthetica, including the Quadra specimen from the Northwest Pacific, were 100% identical to each other. It is notable that COI sequences identified as ‘Myxicola infundibulum’ are divergent and not shown to be monophyletic. Instead, all of the sequences from ‘M. infundibulum’ collected in the Pacific Ocean grouped together with M. aesthetica and Myxicola sp. Quadra, which were also obtained from the Pacific Ocean, rather than with the ‘M. infundibulum’ sequences collected from the Atlantic Ocean (Fig. 7).

Figure 7. Maximum likelihood (ML) tree of slime feather duster worms (Myxicola) including all available COI sequences of the genus available in GenBank. This tree was inferred from 23 COI sequences of 685 sites using RAxML. Posterior probabilities (PP) and ML bootstrap supports (BS) are shown. Sequences obtained in this study from slime feather duster worms are highlighted in bold blue font. The COI sequence of Myxicola aesthetica obtained in this study is grouped with other sequences of M. aesthetica that were also collected from Northeast Pacific Ocean. COI sequences from specimens identified as ‘M. infundibulum’ do not form a monophyletic group, suggesting the presence of cryptic species. Myxicola sp. Quadra forms a distinct clade along with specimens of ‘M. infundibulum’ collected from Bamfield (Canada), indicating that these likely represent a new species that can be distinguished from other M. infundibulum-like host species.
Discussion
1, 2 or 3 species of Selenidium?
Gregarines are highly diverse at both morphological and molecular phylogenetic levels (Leander, Reference Leander2008; Diakin et al., Reference Diakin, Paskerova, Simdyanov, Aleoshin and Valigurová2016; Schrével et al., Reference Schrével, Valigurová, Prensier, Chambouvet, Florent and Guillou2016; Rueckert and Horák, Reference Rueckert and Horák2017; Mathur et al., Reference Mathur, Kolísko, Hehenberger, Irwin, Leander, Kristmundsson, Freeman and Keeling2019; Paskerova et al., Reference Paskerova, Miroliubova, Valigurová, Janouskovec, Kováciková, Diakin, Sokolova, Mikhailov, Aleoshin and Simdyanov2021). When multiple closely related species, especially congeneric species, are found in the same host species or even within one individual, distinguishing them requires careful examination, as different morphotypes might reflect different environmental conditions or developmental stages of the same species (Rueckert et al., Reference Rueckert, Villette and Leander2011).
Based on our examination of morphology, the two morphotypes of Selenidium trophozoites observed in Myxicola sp. Quadra were different in several aspects: infection location within the host, location of the nucleus within the trophozoite, trophozoite length and the presence or absence of cytoplasmic transverse lines visible under the light microscope (Table 2). Although detailed surface features of these two morphotypes examined with SEM did not reveal any noticeable differences, except for their cell lengths (Fig. 3), the pairwise distance and phylogenetic positions inferred from SSU rDNA sequences demonstrated that the two morphotypes represent different species (Table 1 and Fig. 5). The long morphotype of Selenidium from both Myxicola sp. Quadra and M. aesthetica were indistinguishable with SEM (Fig. 3A and 3B), which is consistent with their highly similar (99%) SSU rDNA sequences (Table 1 and Fig. 5). Therefore, we identified two Selenidium species in this study: one (the long morphotype = S. elongatum n. sp.) that infected both Myxicola sp. Quadra and M. aesthetica, and one (the short morphotype = S. cf. mesnili) found only in Myxicola sp. Quadra.
Table 2. Comparison of traits in the Selenidium species inhabiting Myxicola host species
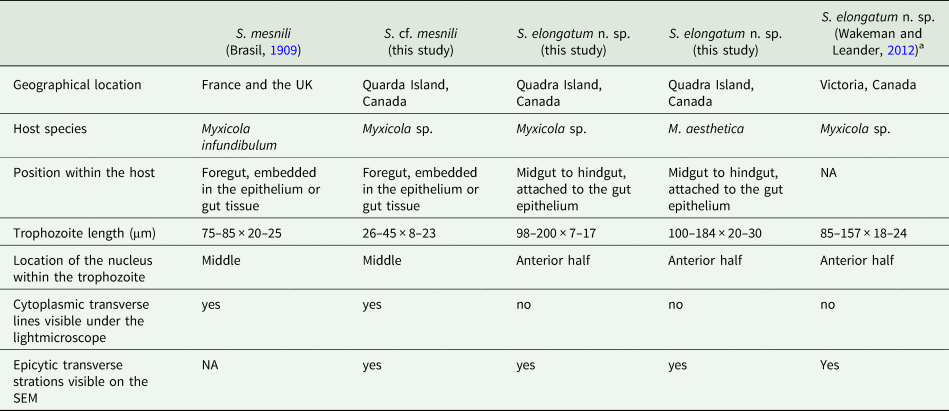
a Reported as S. cf. mesnili in Wakeman and Leander (Reference Wakeman and Leander2012).
Comparison of the 2 morphotypes of Selenidium with S. mesnili
Several authors have studied Selenidium mesnili Brasil, Reference Brasil1909, which infects Myxicola infundibulum originally collected in Europe (France and the UK) in the early 1900s (Brasil, Reference Brasil1909; Ray, Reference Ray1930; Reed, Reference Reed1933). Based on morphological characters, the short morphotype infecting Myxicola sp. Quadra highly resembles S. mesnili described in Europe: its location in the host, cell length, the position of the nucleus and the presence of cytoplasmic transverse lines. Trophozoites of S. mesnili described by Brasil (Reference Brasil1909) measured 75–85 μm long and 20–25 μm wide and were found in the host's epithelium. He mentioned the ‘zebroid’ nature of the cytoplasm ( = cytoplasmic transverse lines) and the presence of ‘grains' near the anterior end of the living cell (see Fig. 2D and 2F). Ray (Reference Ray1930) also noted these characteristics, along with their limited distribution in the anterior part of the gut, and that they spend most of their life cycle within host epithelial cells. The cell length of S. mesnili reported by Ray (Reference Ray1930) was 50–90 μm, with a width of 20–30 μm. Therefore, when comparing the short and long morphotypes observed in this study with S. mesnili from Europe, the short morphotype is more similar to S. mesnili based on the aforementioned characteristics.
Although all these host specimens were identified as ‘Myxicola infundibulum’, it is likely that Myxicola infundibulum actually consists of multiple cryptic species with subtle morphological differences and different geographical ranges (Putignano et al., Reference Putignano, Gravili and Giangrande2023). Unfortunately, molecular phylogenetic data from Myxicola infundibulum and S. mesnili collected from the type location in Europe are not available. However, considering that none of the currently recognized species of Myxicola have the same number of radiole pairs (one of the important taxonomic traits for the group) as Myxicola sp. Quadra (i.e. 21), it is likely that this host species is a new species that differs from M. infundibulum sensu stricto from the type location in Europe, which has 37 radiole pairs (Putignano et al., Reference Putignano, Gravili and Giangrande2023). Consequently, the European hosts from which the trophozoites of S. mesnili were collected, namely M. infundibulum, likely represent a different species from the host Myxicola sp. we collected on Quadra Island, Canada. Despite their significant morphological similarities, without molecular data of the type species, we cannot confirm that the short morphotype Selenidium is indeed S. mesnili. Therefore, we identify the short morphotype Selenidium in Myxicola sp. Quadra as Selenidium cf. mesnili.
The SSU rDNA sequences of the long morphotype of Selenidium from the two Myxicola host species obtained from Quadra Island were 97.8% and 98.6% identical to the SSU rDNA sequence from ‘Selenidium cf. mesnili’ obtained from Myxicola sp. collected in Victoria, Canada (GenBank ID: JN857968), which also exhibited the long morphotype, with a length of 85–159 μm and a width of 18–24 μm (Wakeman and Leander, Reference Wakeman and Leander2012). The clade containing these sequences was strongly supported in our phylogenetic analyses (BS = 99 and PP = 1) and was shown as sister to the short morphotype (Fig. 5). Therefore, we propose a new species of Selenidium for the long morphotype (obtained from Myxicola sp. Quadra, M. aesthetica and Myxicola sp. Victoria) that is clearly distinguished from S. mesnili: Selenidium elongatum n. sp.
Astome ciliates
Understanding the evolutionary relationships among mouthless ciliates has been a challenge due to the importance of oral structures in ciliate taxonomy (Lynn, Reference Lynn2008). Molecular data has proven invaluable for elucidating the phylogenetic placement and relationships among these morphologically streamlined ciliates (Fokam et al., Reference Fokam, Ngassam, Strüder-Kypke and Lynn2011; Sauvadet et al., Reference Sauvadet, Lynn, Roussel, Le Panse, Bigeard, Schrével and Guillou2017; Obert and Vďačný, Reference Obert and Vďačný2019). In fact, recent studies with molecular data suggested that loss of mouth occurred several times independently among oligohymenophoreans (Rataj and Vďačný, Reference Rataj and Vďačný2018; Zhang and Vd'ačný, Reference Zhang and Vd'ačný2022). Although ‘Astomatia’ is polyphyletic, SSU rDNA sequences indicate that astomes infecting annelid hosts (polychaetes and oligochaetes) form a monophyletic group (Fig. 6), consistent with findings from other studies (Sauvadet et al., Reference Sauvadet, Lynn, Roussel, Le Panse, Bigeard, Schrével and Guillou2017; Zhang and Vd'ačný, Reference Zhang and Vd'ačný2022). However, SSU rDNA sequences for astomes infecting polychaete hosts are only available for one genus, namely Durchoniella, thus far.
Polychaete-infecting astomes are classified into 7 genera in three families: Buetschliellidae (Anoplophryopsis, Butschliella, Herpinella and Rhizocaryum), Intoshellinidae (Spirobuetschliella) and Radiophryidae (Durchoniella and Hovasseiella) (Caullery and Mesnil, Reference Caullery and Mesnil1907; Awerinzew, Reference Awerinzew1908; De Puytorac, Reference De Puytorac1954). Although astomes have been identified in several different families of polychaetes (e.g. Eunicidae, Spionidae, Ophelidae, Cirratulidae and Serpulidae), they have never been reported in hosts within the Sabellidae (e.g. Myxicola) (De Puytorac, Reference De Puytorac1954). The family Buetschliellidae is one of the 3 families that include species lacking attachment apparatus, and all the four genera within Buetschliellidae are from different families of polychaete hosts. Species within Buetschliellidae do not have attachment apparatus, such as a hook or a sucker, but do have a bare area without cilia near the anterior end, except for Rhizocaryum. Although Rhizocaryum is also completely covered by cilia, its size is considerably smaller (average of 60 μm long) compared to the astome ciliate we found.
Therefore, the astome ciliate observed in this study represents a new host range for the group, and the absence of an attachment apparatus or a ‘thigmotactic area’ at the anterior end is distinct from other known astomes found in marine annelid hosts. We establish a new genus within the family Buetschliellidae to accommodate the astome ciliate observed in this study (Figs 4 and 6). However, it should be noted that family-level classification of astomes has been chaotic, as gain or loss of organelles appear to have occurred multiple times due to adaptive radiation and host-driven diversification (Obert and Vďačný, Reference Obert and Vďačný2021). An improved phylogeny, including taxa from diverse host groups and environments, would be necessary to clarify the evolutionary relationships among astomes.
Coinfection, host specificity and niche partitioning
S. elongatum n. sp. (long morphotype) was found in both Myxicola species. However, S. cf. mesnili (short morphotype) and Pennarella elegantia n. gen. et sp. (astome ciliates) have never been observed in M. aesthetica and Myxicola sp. Quadra, respectively. This suggests that S. cf. mesnili (short morphotype) and the species of astome ciliates observed here are host specific. The two Myxicola species were collected from different types of habitats; Myxicola sp. Quadra is benthic, while M. aesthetica is typically found higher up in shallower waters (i.e. attached to suspended structures like submerged ropes). The distance between the two collection sites was ~10–20 m. Given the relatively short distance between these two sites and the fact that S. elongatum n. sp. (long morphotype) was shared by both host species, it seems likely that the cysts of these endosymbionts are present in both habitats. However, our understanding of their life cycles and cyst dispersal remains limited. If S. cf. mesnili (short morphotype) and P. elegantia exclusively infect Myxicola sp. Quadra and M. aesthetica, respectively, then this might be attributed to either specific host-endosymbiont cell recognition or suitable environment conditions for these particular symbionts. For instance, in both gregarines and astomes, previous studies have suggested that the localization of different endosymbiont species within the same host individual is correlated with factors such as ionic concentrations (e.g. pH) (Göhre, Reference Göhre1943; Nana and Zéphyrin, Reference Nana and Zéphyrin2014). The fact that P. elegantia was not limited to certain areas of the host gut and was freely swimming in the host gut lumen could be explained by the lack of an attachment apparatus or competition for resources with other coinfecting endosymbiont species (i.e. Selenidium).
While coinfection by multiple gregarines in a single host is not uncommon, there is limited knowledge regarding the specific locations of these symbionts within the host's body (Coulon and Jangoux, Reference Coulon and Jangoux1986; Clopton et al., Reference Clopton, Janovy and Percival1992; Wakeman et al., Reference Wakeman, Heintzelman and Leander2014). Three congeneric gregarine species (Gregarina niphandrodes, G. cuneata and G. polymorpha) occupying different regions of the gut or found at different life stages was seen in Tenebrio molitor, yellow-mealworm beetles (Göhre, Reference Göhre1943; Clopton, Reference Clopton2009). Our study presents the first instance of sister gregarine species occupying distinct regions of the gut in the marine hosts. The mechanisms behind a symbiont preferences for certain cells, tissues and hosts remain unclear. Improved understanding of this study system might help explain interactions between hosts and symbionts, interactions among different symbionts, as well as the environmental factors influencing these interactions.
Taxonomic summary
Apicomplexa Levine, 1970
Archigregarinorida Grasse, 1953
Selenidiidae Brasil, 1907
Selenidium Giard, 1884
Selenidium elongatum n. sp. Park and Leander
Diagnosis. Trophozoite lengths vary, with some reaching considerable lengths (ranging from 50 to 180 × 15–25 μm). The nucleus is ellipsoid in shape and is positioned between the mid and the anterior end of the cell. The mucron can be either round or pointed and is free of folds. The cell surface displays 20–28 epicytic folds, and transverse striations can be visible in the anterior to middle part of the cell. These cells are capable of bending and twisting.
DNA sequence. SSU rDNA sequence has been deposited in GenBank (accession ID: PP109352)
Type locality. Hyacinthe Bay, Quadra Island, British Columbia, Canada (50°6′53″N, 125°13′29″W)
Type habitat. Marine
Type host. Myxicola sp. Quadra (accession ID for the host COI: PP111331)
Location in host. Mid to posterior region of the intestine
Iconotype. Figure 1K.
Zoobank Registration LSID. urn:lsid:zoobank.org:act:D5A5A5EB-87D4-43B1-AF89-212A81F4FEFB
Etymology. The trivial name ‘elongatum’ refers to the elongated shape of the trophozoites.
Remarks. This species was also found in the intestines of M. aesthetica.
Ciliophora Doflein, 1901
Oligohymenophorea de Puytorac et al., 1974
Astomatia Schewiakoff, 1896
Astomatida Schewiakoff, 1896
Buetschliellidae de Puytorac in Corliss, 1979
Pennarella n. gen. Park and Leander
Diagnosis. The elongated cells measure 200–400 μm long and 50–90 μm wide, and they are completely covered by cilia without any attachment apparatus or thigmotactic area. A macronucleus is clearly visible, extending almost the entire length of the cell. These cells swim freely in the gut lumen.
Type species. Pennarella elegantia n. sp. Park and Leander
Etymology. ‘Penn’, a Latin word meaning ‘feather’, refers to the host family ‘Sabellidae’, commonly known as ‘feather duster worms.’
Pennarella elegantia n. sp. Park and Leander
Diagnosis. The elongated cells measure 267 μm long (217–387 μm, n = 6) and 70 μm wide (50–86 μm, n = 6). A clearly visible macronucleus is 207 μm long (150–320 μm, n = 4) and 21 μm wide (20–25 μm, n = 4). A micronucleus may be absent. There are between 6 and 12 contractile vacuoles in a single row, although a second row can be present with a smaller number of vacuoles (1–3). The cell is entirely covered by cilia and lacks an attachment apparatus such as a hook or an apparent thigmotactic area. These cells swim freely within the host's gut lumen.
DNA sequence. SSU rDNA sequence has been deposited in GenBank (accession ID: PP109353)
Type locality. Hyacinthe Bay, Quadra Island, British Columbia, Canada (50°6′53″N, 125°13′29″W)
Type habitat. Marine
Type host. Myxicola aesthetica (accession ID for the host COI: PP111332)
Location in host. Gastrointestinal track of the host
Iconotype. Figure 4A
Zoobank Registration LSID. urn:lsid:zoobank.org:act:0FB773B2-BF65-4D92-94C9-550DAA0D4B69
Etymology. ‘elegantia’ refers to the elegant swimming movement of the ciliates.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024000209.
Data availability statement
All the sequences generated in this study are deposited in GenBank (Accession ID: PP109350-PP109353, PP111331-PP111332). LSID for this publication is urn:lsid:zoobank.org:pub:AC7EECEA-B5FD-42ED-AAB6-7888BA7EE125.
Acknowledgements
We are grateful to Alana Closs, Kyle Hall, Ondine Pontier, Danja Currie-Olsen and Tyrel Froese for their assistance in the Quadra lab and for their contributions to collecting animal specimens through scuba diving. We thank Niels Van Steenkiste and Regine Claire Manglicmot for their help in the field. Also, we would like to thank Derrick Horne for his technical assistance with SEM, and Vittorio Boscaro for sharing his knowledge of ciliates.
Author contributions
EP and BSL conceived and designed the study. EP conducted data gathering and analyses. EP produced the figures and wrote the manuscript with input from BSL.
Financial support
This work was funded by grants to BSL from the Hakai Institute and the National Sciences and Engineering Research Council of Canada (NSERC 2019-03986).
Competing interests
The authors declare there are no conflicts of interest
Ethical standards
Not applicable.