Introduction
Oxyspirura petrowi (Spirurida: Thelaziidae) is a heteroxenous nematode found in a variety of avian species in the USA. O. petrowi infects the eyes of its hosts, situating on the surface of the eye, underneath the nictitating membrane and eyelids, as well as in the ducts and glands behind the eye (Dunham et al. Reference Dunham, Peper, Baxter and Kendall2014a , Reference Dunham, Soliz, Fedynich, Rollins and Kendall b ). First identified in Germany in the family Laniidae (Skrjabin, Reference Skrjabin1929), O. petrowi has since been identified in several other orders of birds including Galliformes and Passeriformes in Michigan (Cram, Reference Cram1937) as well as various parts of the USA since (Saunders, Reference Saunders1935; McClure, Reference McClure1949; Pence, Reference Pence1972; Dunham and Kendall, Reference Dunham and Kendall2017).
Of the regions that O. petrowi has been identified, the Rolling Plains ecoregion of west Texas is one of the most targeted areas of research on this parasite. This is largely because of the decline in northern bobwhite (Colinus virginianus; hereafter bobwhite) within this region. A highly popular gamebird in the USA, bobwhites in the rolling plains have experienced an annual decline of >4% over the past several decades (Sauer et al. Reference Sauer, Hines, Fallon, Pardieck, Ziolkowski and Link2013). The decline has been credited to many factors including habitat loss, habitat fragmentation, agricultural practices and weather conditions (Brennan, Reference Brennan1991; Rollins, Reference Rollins and Brennan2007; Hernandez et al. Reference Hernandez, Brennan, DeMaso, Sand and Wester2013). However, until recently, parasites have remained undervalued in their potential effects on the decline.
Impacts of eyeworm infection in quail was first speculated by Jackson and Galley (Reference Jackson and Galley1963) in Rolling Plains for Oxyspirura sigmoides (=O. petrowi). In his findings, Jackson reported potential damage to the eyes of the bobwhite containing more than 15 eyeworms (Jackson and Green, Reference Jackson and Green1964), as well as strange behaviour that was suspected to be a result of vision impairment (Jackson and Galley, Reference Jackson and Galley1963). Further analysis by Dunham et al. (Reference Dunham, Bruno, Almas, Rollins, Fedynich, Presley and Kendall2016) found lesions and adenitis in the Harderian gland, a gland associated with immune defense (Payne, Reference Payne1994), and corneal scaring in bobwhites infected with O. petrowi. It is likely that the damage caused by these worm burdens can result in reduced foraging efficiency, an inability to effectively escape predators, as well as an inability to avoid stationary objects like a fence or building (Dunham et al. Reference Dunham, Bruno, Almas, Rollins, Fedynich, Presley and Kendall2016).
Despite the increased interest in recent years, O. petrowi’s evolutionary relationships with other parasites are still relatively unexplored. Phylogenetic studies of eyeworms in both humans and animals could be useful in understanding epidemiological, ecological and evolutionary influences on their hosts. A previous phylogenetic analysis using the 18S gene region of O. petrowi showed filarial nematode families to have a close genetic relation to O. petrowi (Xiang et al. Reference Xiang, Guo, Zhang, LaCoste, Rollins, Bruno, Fedynich and Zhu2013). However, Xiang et al. (Reference Xiang, Guo, Zhang, LaCoste, Rollins, Bruno, Fedynich and Zhu2013) suggest that their results are not strongly reliable for the evolutionary affinity of Oxyspirura with other parasites due to lack of sequences in the Thelazioidea super family. This issue can be addressed by constructing multiple phylogenetic trees using different gene regions.
To construct representative and reliable phylogenetic relationships, selecting the appropriate gene regions for analysis is the most important step. Hwang and Kim (Reference Hwang and Kim1999) suggest an improper selection of a gene region can lead to poor understanding of the evolutionary relationship. For this reason, they also note that highly conserved markers of nuclear DNA (rDNA) and hyper variable regions of mitochondrial DNA (mtDNA) have been identified as useful in investigating phylogenetic relationships of higher categorical levels (deep branches) and lower categorical levels (recently diverged branches) of taxonomy, respectively.
The 18S ribosomal subunit (SSU) of nuclear rDNA is suitable due to its highly conserved region for strong evolutionary links as compared with 28S or LSU, 5.8, and Internal Spacers (ITS1 & ITS2) (Hwang and Kim, Reference Hwang and Kim1999). Additionally, 18S has been completely characterized of its V1–V9 variable regions with V4 as the region representing 18S variability in eukaryotes (Nickrent and Sargent, Reference Nickrent and Sargent1991). The V4 region is significant and allows us to distinguish between family and genera and even species in nematode diversity studies. For these reasons, 18S is one of the most popular genetic markers for phylogenetic studies in eukaryotes.
Similarly, mitochondrial DNA has also been used as a popular molecular marker in genetic diversity studies for nearly three decades. In recent years, hundreds of complete parasite mitochondrial genomes have been studied and characterized (Hu and Gasser, Reference Hu and Gasser2006). Among the mitochondrial genes, cytochrome oxidase I (COXI) is preferred as a standardized tool for molecular taxonomy and identification of species (Ratnasingham and Hebert, Reference Ratnasingham and Hebert2007). This gene region is often used as a marker for phylogenetic studies because of its strongly conserved region across species, easiness to amplify in polymerase chain reaction (PCR), lack of introns, lack of recombination, and very small intergenic regions (Galtier et al. Reference Galtier, Nabholz, Glémin and Hurst2009). It is also an efficient tool used for DNA barcoding and nematode identification on species level (Derycke et al. Reference Derycke, Vanaverbeke, Rigaux, Backeljau and Moens2010).
In order to provide more detail to the evolutionary relationships of O. petrowi to other parasites, as previously done with 18S, we use a near-complete 18S and a partial COX1 gene sequence of O. petrowi to generate phylogenetic trees. Presently, there are no phylogenetic studies reported on O. petrowi’s mitochondrial COX1 gene. In this study, we analyze the phylogenetic relationships with the Maximum Likelihood (ML) and Maximum Parsimony (MP) method using MEGA 7 software. By combining the analyses of both 18S and COX1, these results could be useful in understanding O. petrowi’s relationship to other eyeworms as well as its potential effects on the bobwhite based on these evolutionary relationships.
Materials and methods
Ethics statement
This experiment was approved by Texas Tech University Animal Care and Use Committee under protocol 16071-08. All bobwhites were trapped and handled according to Texas Parks and Wildlife permit SRP-0715-095.
Data availability statement
All data generated or analyzed during this study are included in this paper. Sequencing data obtained from this study has been submitted to DNA Data Bank of Japan (DDBJ) (Acc No. LC316613 and LC333364).
Study area
The experimental study area of the present paper is consistent with the study area described in Dunham et al. (Reference Dunham, Soliz, Fedynich, Rollins and Kendall2014b ). The broader range of application (e.g., Rolling Plains) was described by Rollins (Reference Rollins and Brennan2007).
Sample collection
Wild bobwhites were collected in July of 2017 from the same study area, in the same manner and using the same techniques previously described by Dunham et al. (Reference Dunham, Soliz, Fedynich, Rollins and Kendall2014b ). O. petrowi were collected, aged and sexed as previously described by Dunham et al. (Reference Dunham, Soliz, Fedynich, Rollins and Kendall2014b ). Adult eyeworm were washed repeatedly with 1X Phosphate-buffered Saline (PBS). Samples were preserved in 95% ethanol and stored at −80 °C until DNA extraction.
DNA extraction
Genomic DNA of adult O. petrowi was extracted using Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer's instruction with slight modifications. Modifications included homogenization of eyeworms in 180 µL of ATL buffer with a micro-pestle (Sigma, USA) followed by an addition of 20 µL proteinase K. Additionally, samples were incubated at 56 °C for 20 min and an elution of 100 µL sterile water was performed as the final step. Extracted DNA was stored at −20 °C until further use.
Primer designing
Primers for 18S were designed based on CLUSTAL W2 multiple sequence alignment results. Two forward primers and a reverse primer for 18S were designed and validated using online primer designing tools (Table 1). An internal primer was designed based on the sequence results of 18S to obtain an internal sequence. For COX1, initial amplification was done using degenerative nematode-specific primers (Prosser et al. Reference Prosser, Velarde-Aguilar, Leon-Regagnon and Hebert2013) and primers were designed (Table 1) using sequencing results of primary amplification based on methods described in Kalyanasundaram et al. (Reference Kalyanasundaram, Blanchard and Kendall2017).
Table 1. Oligonucleotide primers for amplifying and sequencing 18S and COX1 regions of O. petrowi

a Used to obtain missing 18S region by sequencing.
Amplification of O. petrowi 18S and COX1
Both sets of primers were optimized using an annealing temperature gradient from 55 to 60 °C. PCR reactions contained 5 µL of 2X Red Dye Master Mix (Bioline, England), 0.5 µL of 10 µ m forward and reverse 18S and COX1 primers, 3.0 µL of molecular grade water, and 1 µL of O. petrowi template DNA for a total reaction volume of 10 µL for 18S and COX1, respectively. PCR reactions were run under the following parameters: 95 °C for 3 min, 95 °C for 30 s, 57 and 60 °C for 30 s, for both COX1 and 18S. Elongation temperature was kept at 72 °C for 2 min for 18S reactions and 30 s for COX1 reactions with 29 cycles. Final elongation at 7 °C for 5 min was used to check extended chain. Amplification of the 18S and COX1 products were visualized on 1.5% agarose gels.
Sequencing
Purified PCR products of 18S and COX1 reactions were sequenced in both directions using their respective forward and reverse primers. Based on the sequencing results of 18S, an internal primer was designed and used to amplify 18S in PCR again. This PCR product was then sequenced to obtain the near complete 18S rDNA sequence. The partial COX1 sequence was confirmed with similar methods as described by Kalyanasundaram et al. (Reference Kalyanasundaram, Blanchard and Kendall2017) using Thelazia callipaeda COX1 sequence (Liu et al. Reference Liu, Gasser, Otranto, Xu, Shen, Mohandas, Zhou and Zhu2013) as a comparison. Raw sequences were trimmed using DNA chromatogram explorer (www.dnabaser.com). Final sequences used for analysis totaled at 1811 bp for 18S and 598 bp for COX1. Sequence similarity was performed using BLAST analyses.
Phylogenetic analysis
MEGA 7 software was used to generate phylogenies of 18S and COX1 gene regions. O. petrowi 18S and COX1 were separately aligned with selected sequences of other parasites from the GenBank, NCBI. Initially, we did multiple alignments with nearly 150 sequences of 18S retrieved from GenBank using CLUSTAL W program and simple trees were constructed by ML method (Larkin et al. Reference Larkin, Blackshields, Brown, Chenna, McGettigan, McWilliam, Valentin, Wallace, Wilm, Lopez, Thompson, Gibson and Higgins2007). We used taxa from order Spirurida for constructing 18S phylogenetic tree. Based on the alignment results, identical and unfit/short sequences were removed until enough quality congregate sequences were made. All gaps were removed and the total 1634 positions were used in the final dataset. Similarly, we constructed phylogenetic tree for COX1 sequences retrieved from GenBank. We used Filarioidea and Thelazioidea super family as major taxa to generate COX1 phylogeny both ML and MP method. Species specifically analyzed in this study include Brugia malayi, Wuchereia bancrofti, Loa loa, Dirofilaria repens from Filarioidea and O. petrowi, T. callipaeda from Thelazioidea super family. The complete deletion was used to treat gaps as missing information and totalled 405 positions in the final dataset. Phylogenetic tree constructions were performed using character state including ML and MP method. The bootstrap value was set at 1000 in order to represent strong evolutionary relationships between O. petrowi and other parasites of the Nematoda phylum.
Results
BLAST analysis results of the 1811 bp sequence of 18S showed a 100% identity to O. petrowi isolates (KF110800-KF110799), confirming that our sequence corresponds with previously submitted sequences of O. petrowi. Sequence results of O. petrowi’s 18S gene region revealed a 95% to 96% similarity to the 18S region of B. malayi (AF036588), W. bancrofti (LM006781), L. loa (XR-002251421) and a 92% similarity to T. callipaeda (LK982445). Oxyspirura petrowi’s COX1 shows an 86% similarity to Dirofilaria spp. (KX265050) and an 85% similarity to Dirofilaria repens (KX265049). Lastly, there is also an 84% similarity between O. petrowi's and T. callipaeda’s COX1 gene region (KY908318-KY908318). Both O. petrowi’s 1811 bp 18S sequence (Fig. 1A) and 598 bp COX1 sequence (Fig. 1B) were submitted in DDBJ (Acc No. LC316613 and LC333364).
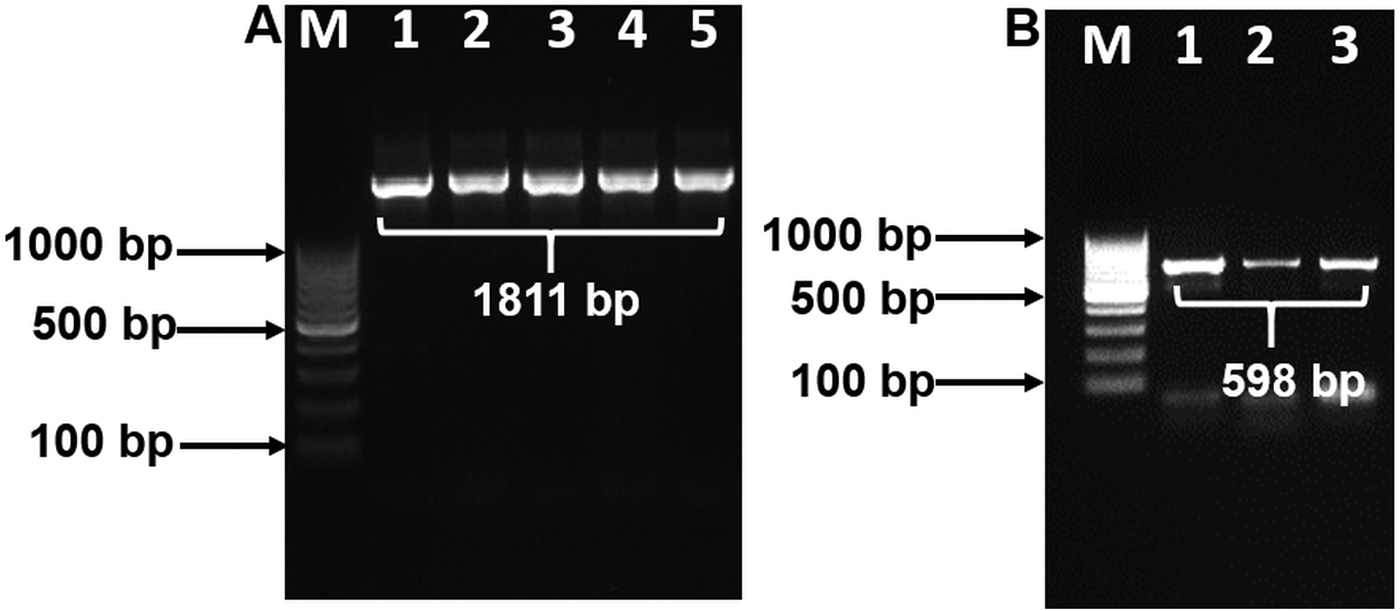
Fig. 1. Polymerase chain reaction (PCR) amplification of 18S and COX1 gene using specific primers. (A) 18S rDNA amplification Lane M: 100 bp DNA ladder (Fermentas); Lane 1–5: 18S rDNA amplicon (1811 bp). (B) partial COX1 gene amplification. Lane M: 100 bp DNA Marker (Fermentas); Lane 1–3: partial COX1 amplified products (598 bp).
A phylogenetic tree was constructed for both 18S and COX1 gene regions of O. petrowi to determine its evolutionary relationship within the Nematoda phylum (Figs 2 and 3). All O. petrowi isolates from different geographical locations were placed in one cluster and received strong support by ML and MP bootstrap analysis (100%). All clades in the 18S tree received moderate to high (50–100%) support by ML bootstrap analyses. Bootstrap values below 50% were removed from the COX1 trees (Fig. 3A and B). In both 18S and COX1 trees, species of the Filarioidea superfamily placed closely to species of the Thelazioidea superfamily (Figs 2 and 3). T. callipaeda (KY908318) is located within the same clade of O. petrowi in the 18S tree. Similarly, in the COX1 trees, O. petrowi shares a branch of the tree with T. callipaeda. Heliconema longissimum (GQ332423) and Spirocerca spp. (KJ605487) in the Spiroroidea superfamily were also placed in the same clade of the COX1 trees (Fig. 3A and B).
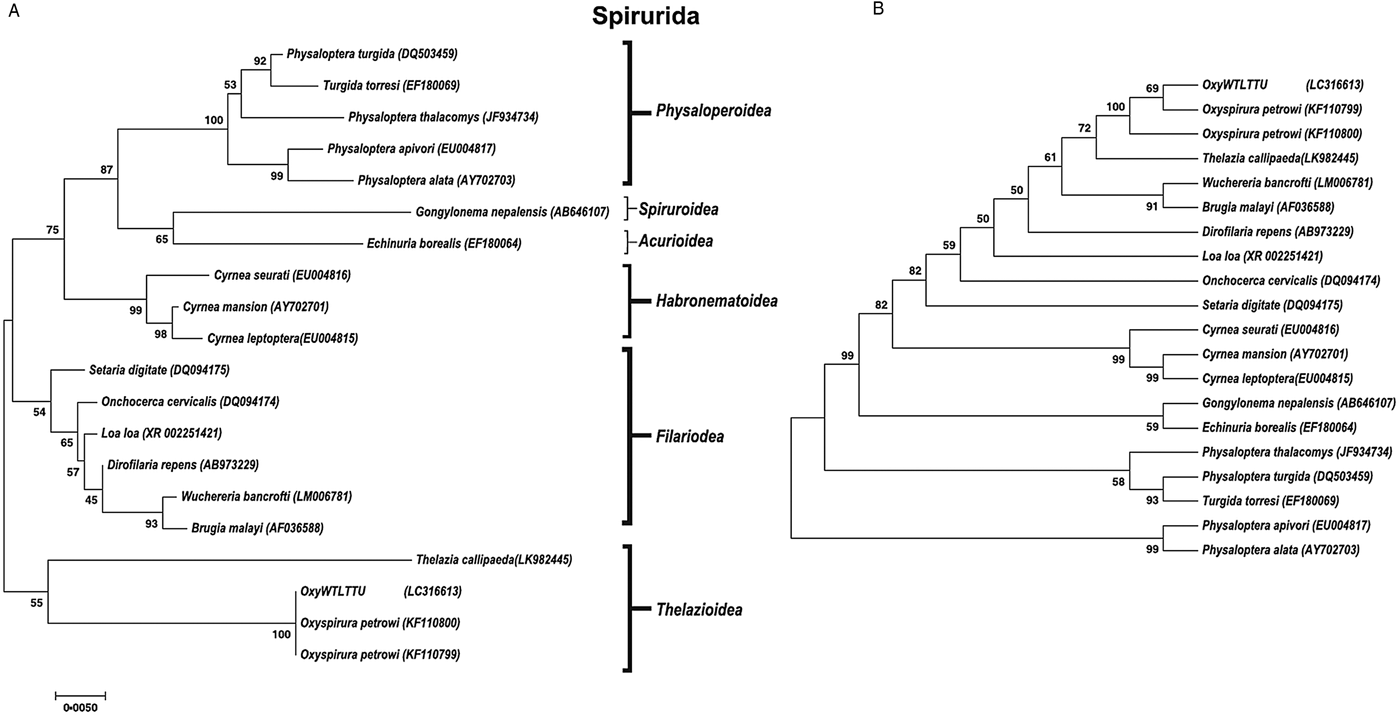
Fig. 2. Phylogenetic analysis of O. petrowi based on a near-complete 18S using Maximum Likelihood and Maximum Parsimony methods. (A) Maximum Likelihood: The evolutionary history was inferred using the ML method based on the Tamura-Nei model. The phylogenetic tree illustrates 18S rDNA sequences of nematodes related to O. petrowi. Bootstrap values above 50 are shown in the tree. All positions containing gaps and missing data were eliminated. Species name and their nucleotide accession numbers were included in the tree. There were a total of 1634 positions for 18S in the final dataset. Evolutionary analyses were conducted in MEGA7. (B) Maximum Parsimony: The evolutionary history was inferred using the MP method based on Subtree-Pruning-Regrafting (SPR) algorithm. The phylogenetic tree illustrates 18S rDNA sequences of nematodes related to the eyeworm O. petrowi.

Fig. 3. Phylogenetic analysis of O. petrowi based on partial COX1 using Maximum Likelihood and Maximum Parsimony methods. (A) Maximum Likelihood: The evolutionary history was inferred using the ML method based on the Tamura-Nei model. The phylogenetic tree illustrates COX1 sequences of nematodes related to O. petrowi. All positions containing gaps and missing data were eliminated. Species names and corresponding nucleotide accession numbers were included in the tree. There was a total of 405 positions for 18S in the final dataset. Evolutionary analyses were conducted in MEGA7. (B) Maximum Parsimony: The evolutionary history was inferred using the MP method based on Subtree-Pruning-Regrafting (SPR) algorithm. The phylogenetic tree illustrates 18S rDNA sequences of nematodes related to O. petrowi.
Discussion
Over the past several decades, molecular phylogenetic studies have received widespread attention in determining evolutionary relationships between various specimens as proposed by Nadler (Reference Nadler1995) in their phylogenetic case study of Ascaridinae nematodes. When morphological features are not similar in parasites, a molecular comparison involving phylogenetic investigation is a useful method to infer the genetic relationship between species (Nadler, Reference Nadler1995). Undoubtedly, morphological evolution can happen strictly on a genetic basis. Comparisons on the genetic level can also decisively confirm or deny relationships between parasites previously examined using morphological characteristics. In this study, we observe the nuclear 18S region and mitochondrial COX1 region of O. petrowi to better understand these relationships not readily available by morphology alone.
Our sequences for 18S and COX1 were confirmed using BLAST analyses. Constructed phylogenetic trees following BLAST analyses for both 18S and COX1 sequences show strong support for the monophyly of the genus Oxyspirura. We also found our phylogenetic results of 18S in congruence with results of Xiang et al. (Reference Xiang, Guo, Zhang, LaCoste, Rollins, Bruno, Fedynich and Zhu2013). Based on 18S results from both studies, all the parasite species within the superfamilies of the phylogenetic trees are within the order Spirurida, and the phylogenetic trees reveal the Oxyspirura genus as a sister group for the Filarioidea superfamily.
Although the various species of Filarioidea identified in this study all have a 96% similarity with O. petrowi’s 18S region, L. loa has lower nucleotide variation, indicating that L. loa is of closer relation to O. petrowi. While the filarial nematodes W. bancrofti and B. pahangi cause lymphatic filariasis in the definitive hosts, L. loa causes loaiasis (Chandy et al. Reference Chandy, Thakur, Singh and Manigauha2011; Tan et al. Reference Tan, Fong, Mahmud, Muslim, Lau and Kamarulzaman2011). Typically found in humans of west and central Africa, L. loa is transmitted through a deerfly (Chrysops spp.) vector, with infective larvae entering the wound produced by the deerfly and maturing in subcutaneous tissue (CDC, 2015). Loiasis is caused by both adult worms and microfilaria with clinical symptoms of eosinophilia, Calabar swelling, and eyeworm migration in its hosts (Antinori et al. Reference Antinori, Schifanella, Million, Galimberti, Ferraris, Mandia, Trabucchi, Cacioppo, Monaco, Tosoni, Brouqui, Gismondo, Giuliani and Corbellino2012). During eye worm migration, the parasite may be seen moving in the vitreous cavity, found in the anterior chamber, or in the cornea, resulting in inflammation and impaired vision (Barua et al. Reference Barua, Barua, Hazarika and Das2005; Nayak et al. Reference Nayak, Sinha and Nayak2016).
Treatment of L. loa can be dangerous as it can cause brain inflammation and sometimes complications such as neuropathy and encephalopathy can occur (CDC, 2015). A recent genomic study on L. loa found several orthologous kinases that can be targeted by drugs currently approved for use in humans such as imatinib (Desjardins et al. Reference Desjardins, Cerqueira, Goldberg, Hotopp, Haas, Zucker, Ribeiro, Saif, Levin, Fan, Zeng, Russ, Wortman, Fink, Birren and Nutman2013), potentially providing a safer treatment. Additionally, Xiang et al. (Reference Xiang, Guo, Zhang, LaCoste, Rollins, Bruno, Fedynich and Zhu2013) suggest that the treatment strategies used for human eyeworm infection such as L. loa would be a model to develop treatment strategies for O. petrowi infection in quail.
An additional filarial nematode, D. repens of the Filarioidea superfamily, was identified with COX1 sequencing results. Similar to the 18S results, the COX1 phylogenetic tree placed the Thelaziidae family close to the Filarioidea superfamily and revealed less divergence to D. repens. These observations signify both are sister groups and likely suggests they evolved from the same ancestor. Dirofilaria spp. are responsible for most filarioid eye infections and for nodules on the orbital zone or eyelid (Otranto and Eberhard, Reference Otranto and Eberhard2011; Mateju et al. Reference Mateju, Chanova, Modry, Mitkova, Hrazdilova, Zampachova and Kolarova2016). Found in humans and other domestic and wild animals, D. repens is transmitted by mosquitos (Czajka et al. Reference Czajka, Becker, Jost, Poppert, Schmidt-Chanasit, Kruger and Tannich2014). Another sister group to the Thelazioidea superfamily, the Spirocercidae family, causes spirocercosis in their definitive Canidae hosts in tropical and subtropical regions. Clinical symptoms of spirocercosis include vomiting, odynophagia, hyper salivation and lesions, with aortic lesions being the most common and can be deadly (Van der Merwe et al. Reference Van der Merwe, Kirberger, Clift, Williams, Keller and Naidoo2008). Van der Merwe et al. (Reference Van der Merwe, Kirberger, Clift, Williams, Keller and Naidoo2008) also states that species in this family utilize dung beetles as the intermediate host.
Furthermore, our findings in the 18S and COX1 phylogenetic trees demonstrate the relation of O. petrowi and T. callipaeda in the Thelaziidae family as they were placed in the same clade of our phylogenetic trees. T. callipaeda is an eyeworm responsible for the neglected tropical disease known as thelaziasis in humans and carnivores of Europe and the East Asia. Thelaziasis produces clinical signs of epiphora, conjunctivitis, and ulcerative keratitis in their hosts (Otranto et al. Reference Otranto, Lia, Buono, Traversa and Giangaspero2004). Additionally, T. callipaeda uses fruit flies, Phortica spp., as the intermediate host to transmit infection (Otranto et al. Reference Otranto, Lia, Buono, Traversa and Giangaspero2004; Otranto and Eberhard, Reference Otranto and Eberhard2011).
Based on these results, it is plausible that O. petrowi could have similar impacts on the bobwhite as these parasites have on their hosts. Future studies need to be carried out on pathological relations between O. petrowi and these parasites to fully understand this comparison and effective treatment strategies. Similarly, all described parasites require an intermediate host to transmit infection. While it is postulated that the plains lubber grasshopper (Brachystola magna) is a potential intermediate host (Kistler et al. Reference Kistler, Hock, Hernout, Brake, Williams, Downing, Dunham, Kumar, Turaga, Parlos and Kendall2016), it has not been determined whether it is capable of transmitting to bobwhite. Future investigations into the intermediate host of O. petrowi should prioritize similar species as the intermediate hosts of the related parasites.
This is the first report of examining a partial sequence of the mitochondrial gene region of O. petrowi as well as the first report in comparing this region with the almost-complete nuclear 18S gene region of O. petrowi with other parasites via phylogenetic analyses. In spite of partial sequences, both 18S and COX1 phylogenetic results strongly concluded the relationship of O. petrowi with Thelaziidae family. However, further sequencing of the entire COX1 gene region will help in better understanding inter- and intra-species similarities. Using phylogenetic networking, the COX1 gene region of O. petrowi could potentially be used as a biological tag to study the bobwhite population decline. Future genetic analyses could also help in further characterizing O. petrowi and how it relates to its contribution to the decline of bobwhites of the Rolling Plains ecoregion of Texas.
Acknowledgements
We thank Rolling Plains Quail Research Foundation (23A470) and Park Cities Quail (24A175) for their continued financial support of our quail research. We thank the owners and employees of our study ranch for allowing access and providing lodging. Thank you to the Wildlife Toxicology Laboratory personnel for their field and laboratory assistance.
Financial support
Funding for this work was provided by Park Cities Quail and Rolling Plains Quail Research Foundation (awarded to Ronald J. Kendall, Ph.D.).
Conflict of interest
None.






