Introduction
Musk deer (genus Moschus, family Moschidae) are endemic to the Palearctic region, inhabiting fragmented areas of the Himalayan mountains, the Tibetan Plateau and the adjoining mountainous region in China and eastern Russia (Pan et al., Reference Pan, Wang, Hu, Sun, Zhu and Meng2015). They are solitary habitat specialists, mostly found in forests, alpine shrubland and above the treeline of alpine meadows. Seven species are recognized, of which five occur in the Himalayan range: the Kashmir M. cupreus, Alpine M. chrysogaster, Himalayan M. leucogaster, forest M. berezovskii and black M. fuscus musk deer (Grubb, Reference Grubb, Wilson and Reeder2005). Their populations are declining because of poaching for their musk pods, and habitat fragmentation and degradation. Because of unsustainable exploitation, all musk deer species have been included in the Appendices of CITES since 1979 (Zhou et al., Reference Zhou, Meng, Feng, Yang, Feng and Xia2004). Six musk deer species are categorized as Endangered on the IUCN Red List, and one as Vulnerable. In India, musk deer are included in Schedule I under the Wild Life (Protection) Act, 1972. Because of overlapping distribution ranges and morphological similarities, there is ambiguity regarding the taxonomy of musk deer species, hindering efficient conservation efforts (Pan et al., Reference Pan, Wang, Hu, Sun, Zhu and Meng2015).
The Kashmir musk deer is a little-studied species that occurs in Afghanistan (Ostrowski et al., Reference Ostrowski, Rahmani, Ali, Ali and Zahler2016), Pakistan, India (Timmins & Duckworth, Reference Timmins, Duckworth and .2008) and Nepal (Singh et al., Reference Singh, Khatiwada, Saud and Jiang2019). The historical distribution of musk deer species in the western Himalayan region has been primarily based on phenotypic characteristics such as external morphology and skull morphometry (Groves et al., Reference Groves, Wang and Grubb1995). Musk deer are cryptic, making species validation based solely on morphological traits unreliable (Grubb, Reference Grubb1982; Groves et al., Reference Groves, Wang and Grubb1995; Su et al., Reference Su, Wang and Wang2001). A recent study using molecular and camera-trap data reported a new record of Kashmir musk deer from Mustang in Nepal, west of the Annapurna Himalayas range (Singh et al., Reference Singh, Khatiwada, Saud and Jiang2019), warranting an assessment of the range of the species. The Himalayan musk deer is morphologically similar to the Kashmir musk deer, with seasonally variable coat colours (Liu & Groves, Reference Liu and Groves2014; Singh et al., Reference Singh, Khatiwada, Saud and Jiang2019). Shukla et al. (Reference Shukla, Joshi, Kumar, Thakur, Mehta, Sathyakumar and Goyal2019) indicated the presence of different ecomorphs in musk deer species from Uttarakhand, India, suggesting the presence of distinct lineages whose validation warrants further investigation with molecular methods.
Advanced molecular tools for species identification and phylogenetic analysis have previously resolved phylogenetic complexities in musk deer and aided species validation (Su et al., Reference Su, Wang and Wang2001; Pan et al., Reference Pan, Wang, Hu, Sun, Zhu and Meng2015). Genetic methods confirmed the presence of Himalayan musk deer, previously misidentified as Alpine musk deer, in Tibet (Guo et al., Reference Guo, Li, Zhang and Chen2019). The mitochondrial DNA (mtDNA) control region has proven to be a robust marker for investigating intra-species genetic variation (Hu et al., Reference Hu, Fang and Wan2006; Peng et al., Reference Peng, Liu, Zou, Zeng and Yue2009; Kumar et al., Reference Kumar, Ghazi, Hussain, Bhatt and Gupta2017; Gupta et al., Reference Gupta, Kumar, Angom, Singh, Ghazi, Tuboi and Hussain2018). Here, we describe novel genetic evidence for the presence of the Kashmir musk deer in the western Himalayan region of Uttarakhand, and assess its phylogenetic relationship with other musk deer species. Our findings provide insight into the range, evolutionary history and phylogeograpy of this enigmatic musk deer.
Methods
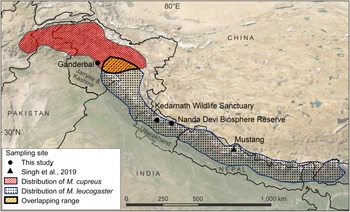
The samples used in this study originated from Kedarnath Wildlife Sanctuary and Nanda Devi Biosphere Reserve, both of which are located in Uttarakhand state, India. The 975 km2 Kedarnath Wildlife Sanctuary is rich in biodiversity and one of the largest protected areas in the western Himalaya, covering altitudes of 1,025–3,662 m. To the east of Kedarnath Wildlife Sanctuary, the Valley of Flowers National Park forms a part of the Nanda Devi Biosphere Reserve (6,407 km2, altitude 1,800–7,817 m). The Ganderbal District of Jammu and Kashmir is a hilly and semi-hilly area of 1,045 km2, at altitudes of 1,650–3,000 m.
Samples and DNA extraction
We used a total of eight tissue samples of musk deer: six from Kedarnath Wildlife Sanctuary, one from Nanda Devi Biosphere Reserve, and one from Ganderbal, Jammu and Kashmir (Fig. 1, Supplementary Table 1). Seven of these samples (MDUK1–MDUK5, MDUK18 and MDJK1) were sent by the State Forest Departments to the Wildlife Forensic laboratory of the Wildlife Institute of India, and sample MDUK19 was collected in Nanda Devi Biosphere Reserve by a team from the Wildlife Institute of India. In addition, we included 12 musk pods sent by the Forest Department from the Chamoli district of the Kedarnath Wildlife Division, to assess the phylogenetic relationship between these samples and the samples from Uttarakhand and Jammu and Kashmir. We extracted genomic DNA (gDNA) from the samples using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) protocol.
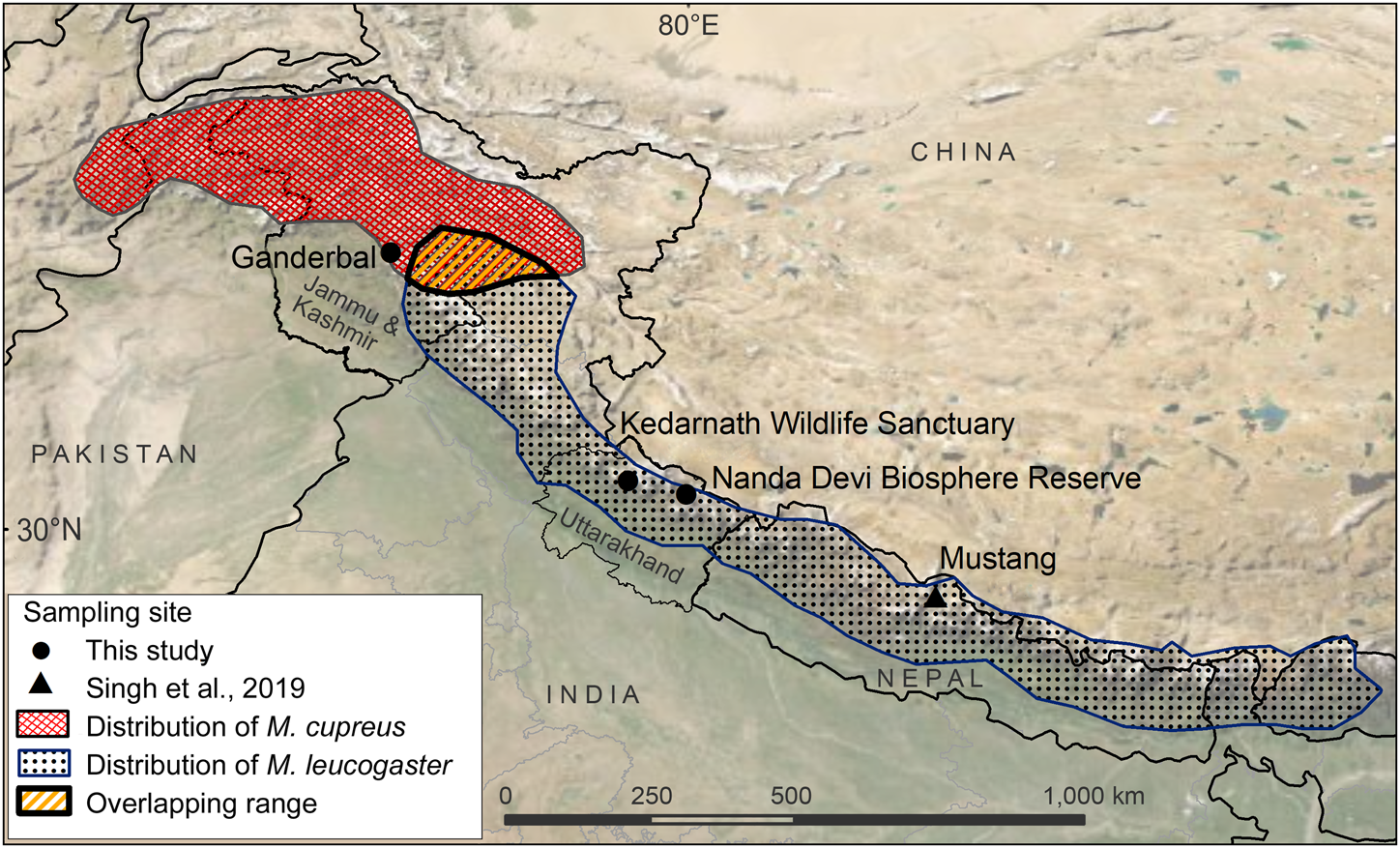
Fig. 1 Sampling locations from this study and Sing et al. (Reference Singh, Khatiwada, Saud and Jiang2019), and distribution ranges of Kashmir musk deer Moschus cupreus and Himalayan musk deer Moschus leucogaster according to the IUCN Red List assessments (Timmins & Duckworth, Reference Timmins and Duckworth2015a,Reference Timmins and Duckworthb).
PCR amplification and sequencing
We performed the PCR in 20 μl reaction volumes containing 10–20 ng of extracted gDNA. The PCR master mix contained: 1 × PCR buffer (Applied Biosystems, Thermo Fisher Scientific, Waltham, USA), 2.0 mM MgCl2, 0.2 mM of each dNTP, 2 pmol of each primer, and 5U of Taq DNA polymerase. We successfully amplified 485-bp-long portions of mtDNA control region using the primers Cerv.tPro: 5′-CCACYATCAACACCCAAAGC-3′ and CervCRH: 5′-GCCCTGAARAAAGAACCAGATG-3′ (Balakrishnan et al., Reference Balakrishnan, Monfort, Gaur, Singh and Sorenson2003). The PCR conditions were: an initial denaturation for 5 minutes at 95 °C, followed by 35 cycles at 95 °C for 45 s, 55 °C for 45 s and 72 °C for 45 s, with a final extension of 72 °C for 15 minutes. The efficiency and reliability of PCR were monitored using positive and negative control reactions. The PCR products were electrophoresed on 2% agarose gel and visualized under UV light. Positive amplicons were treated with Exonuclease-I and USB Shrimp alkaine phosphatase (Affymetrix, Inc., Santa Clara, USA) for 15 min each at 37 °C and 80 °C, respectively, to remove any reaction residues. The purified fragments were sequenced directly in an Applied Biosystems Genetic Analyzer 3500XL from both primers, and set using a BigDye 3.1 kit (Applied Biosystems, Thermo Fisher Scientific, Waltham, USA).
Data analysis
We sequenced the generated amplicons from both directions of targeted mtDNA fragments and edited them with SEQUENCHER 4.9 (Gene Codes Corporation, Ann Arbor, USA). The sequences were aligned and visually inspected using the CLUSTAL X 1.8 multiple alignment programme (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997). We validated the species of all eight samples with known geographical origin (MDUK1–MDUK5, MDUK18, MDUK19 and MDJK1) by comparison with published musk deer sequences: M. moschiferus (n = 2), M. chrysogaster (n = 3), M. anhuiensis (n = 1), M. leucogaster (n = 20), M. berezovskii (n = 24) and M. cupreus (n = 5) from GenBank (National Center for Biotechnology Information, Bethesda, USA; Supplementary Table 2). Because sequences of M. cupreus from Afghanistan were not available in GenBank, we validated M. cupreus using sequences from Nepal (Singh et al., Reference Singh, Khatiwada, Saud and Jiang2019). Thereafter, the sequences of seized musk pods (n = 12) from Uttarakhand were further validated with data from samples of known geographical origin, and with sequences from Singh et al. (Reference Singh, Khatiwada, Saud and Jiang2019). All sequences were further included for phylogenetic analysis.
We calculated the number of haplotypes in the dataset (from the eight samples of known provenance, 12 seized musk pods and M. cupreus sequences from Nepal) using DnaSP 5.0 (Librado & Rozas, Reference Librado and Rozas2009). The Bayesian phylogenetic tree among all the mtDNA control region sequences of musk deer was constructed using BEAUti and the BEAST 1.7 (Drummond et al., Reference Drummond, Suchard, Xie and Rambaut2012). We used a sequence from the Indian mouse deer Moschiola indica (NC037993) as the outgroup. We deployed the best-fit nucleotide substitution model Hasegawa–Kishono–Yano (HKY) + G + I to obtain the best tree topology in phylogenetic analysis. The Markov chain Monte Carlo process was run for 10 million generations with a random starting tree and we sampled one tree every 1,000 generations. We discarded the first 25% of generations as burn-in. Convergence of values and effective sample size was assessed in Tracer 1.7 (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018). The resulting phylogenetic trees were visualized in FigTree 1.4.0 (Rambaut & Drummond, Reference Rambaut and Drummond2012). The spatial distribution of haplotypes was generated using a median-joining network in PopART (Leigh & Bryant, Reference Leigh and Bryant2015). The evolutionary divergence among sequence pairs between musk deer groups was estimated with a p-distance model, including substitution transition and transversion calculated in MEGA X (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018); p-distance is the proportion of nucleotide sites by which two compared sequences differ.
Results
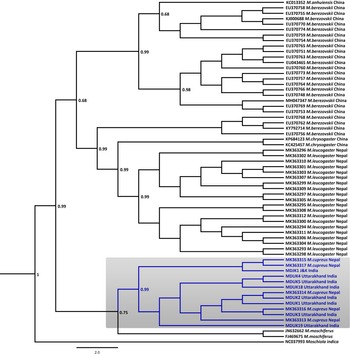
In the Bayesian phylogenetic analysis, the seven samples from Uttarakhand (MDUK1–MDUK5, MDUK18, MDUK19) clustered into a clade with a high posterior probability (PP) value (PP~0.99) with M. cupreus sequences from Jammu and Kashmir (MDJK1) and published sequences from Nepal, whereas M. moschiferus formed the basal clade (PP~0.75; Fig. 2). All seized musk pods (MDUK6–MDUK17) showed 98.5–100% homology with the M. cupreus sequences in GenBank and also clustered within the clade of M. cupreus (Supplementary Fig. 1). All 25 sequences of musk deer from this study and from Singh et al. (Reference Singh, Khatiwada, Saud and Jiang2019) were grouped into eight haplotypes (Supplementary Table 3). Haplotype 1 was common in samples from both Nepal and Uttarakhand, representing three and seven sequences, respectively. Five unique haplotypes (Haplotypes 3−7) were only present in the samples from Uttarakhand, whereas Haplotypes 2 and 8 were unique to samples from Nepal and Jammu and Kashmir, respectively. The Bayesian Inference tree topology indicated that M. cupreus and M. moschiferus had evolved earlier than M. chrysogaster, M. anhuiensis, M. leucogaster and M. berezovskii (Supplementary Fig. 2). We have submitted the newly identified haplotypes of M. cupreus to GenBank (Accession numbers MT822697–MT822703). The sequences of six musk deer species were grouped into 38 haplotypes (Supplementary Fig. 3). The median-joining network showed weak haplotype clustering within M. berezovskii and M. chrysogaster, whereas strong structuring was observed in M. cupreus and M. leucogaster.
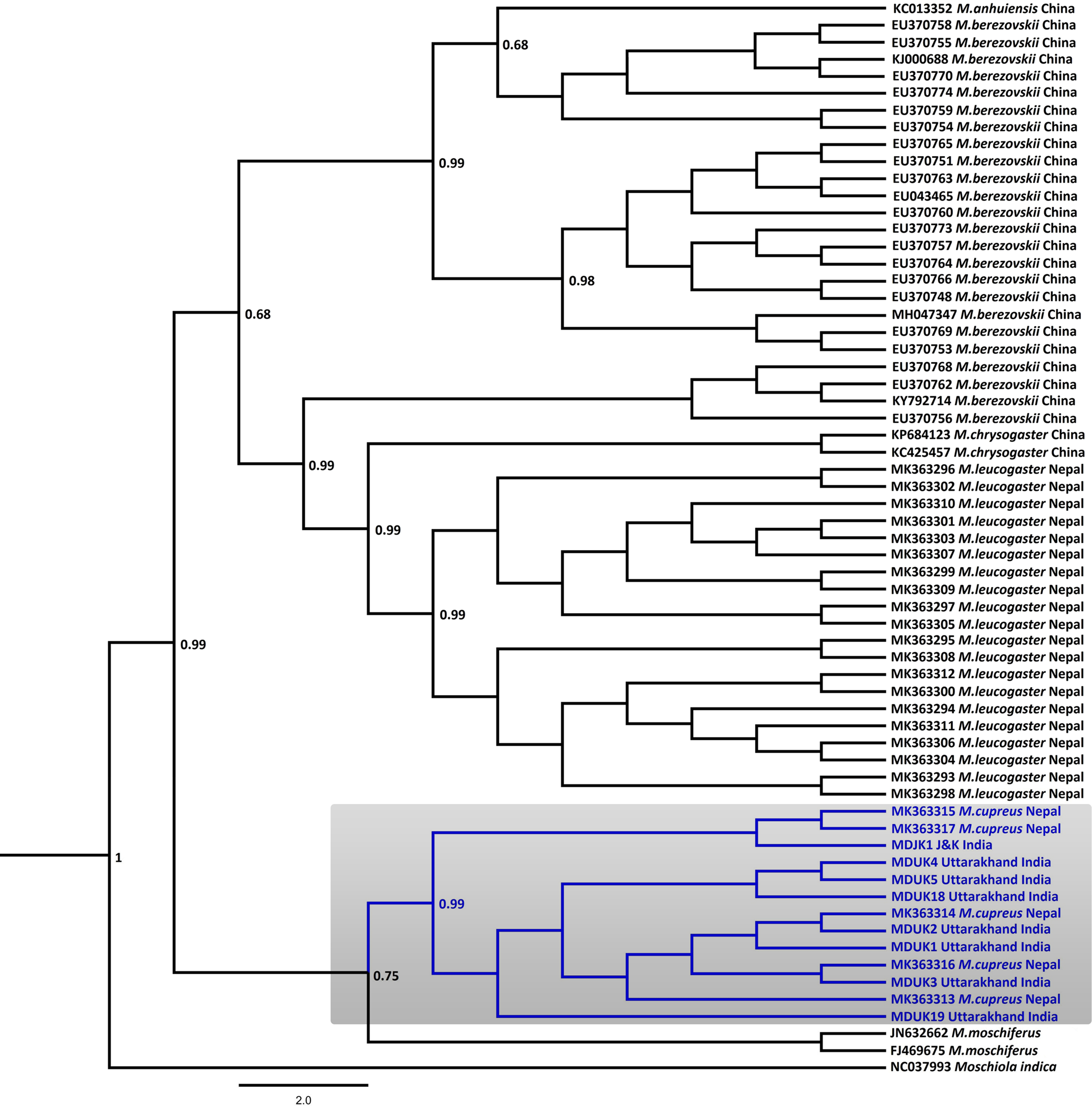
Fig. 2 Bayesian (Markov chain Monte Carlo) consensus tree of musk deer based on the mtDNA control region. Posterior probability values are provided at their respective nodes. The Indian mouse deer Moschiola indica (NC037993) was used as the outgroup. The clade of the Kashmir musk deer M. cupreus is shaded. The scale bar represents substitutions of nucleotides.
The mean pairwise genetic distance analysis indicated that M. cupreus from Nepal were genetically similar to the Jammu and Kashmir and Uttarakhand populations, with low sequence divergences estimated between the groups (1%) and within the species group (0.8%). Among the musk deer species, M. cupreus was closest to M. moschiferus (8.8–9.0%) followed by M. leucogaster (10%), whereas the maximum genetic difference was observed with M. berezovskii (10.9%) (Table 1). High intra-species divergences were observed in M. chrysogaster (6.3%) and M. berezovskii (4.7%). We observed weak genetic clustering within M. berezovskii, forming two separate clades. Moreover, the complete mitogenome sequence (JQ608470) of M. chrysogaster clustered within the M. berezovskii clade, raising concerns over its validity (Supplementary Figs 2 & 3). The high divergence in M. berezovskii requires research to facilitate lineage confirmation.
Table 1 Genetic p-distance of the mtDNA control region of the genus Moschus are represented below the diagonal and standard error values are shown above the diagonal.

Discussion
Our study indicates the presence of M. cupreus in Jammu and Kashmir and Uttarakhand, India. The analysis was supported by strong phylogenetic affinities with recently identified lineages of M. cupreus from Mustang, Nepal (Grubb, Reference Grubb, Wilson and Reeder2005; Singh et al., Reference Singh, Khatiwada, Saud and Jiang2019). It was also corroborated by predictions based on mapping of climate refugia and habitat suitability that suggested the occurrence of M. cupreus in the Himalayan belt stretching from central Nepal to north-west India (including Uttarakhand and Himachal Pradesh), through the Kashmir region to Afghanistan (Singh et al., Reference Singh, Mainali, Jiang, Thapa, Subedi and Awan2020).
The genetic evidence presented here highlights the need for extensive sampling of musk deer species across their range, to facilitate reliable identification of species and to update information on their distribution. Uttarakhand's western Himalayan region is believed to harbour M. leucogaster (Timmins & Duckworth, Reference Timmins and Duckworth2015b) as well as M. cupreus (Timmins & Duckworth, Reference Timmins and Duckworth2015a). However, all samples analysed in this study were of M. cupreus, which raises the possibility that M. cupreus may have been misidentified as M. leucogaster because of morphological similarities between the two species. This warrants a comprehensive reassessment of the distribution ranges of M. cupreus and M. leucogaster in the western Himalayas, to enable effective management of these threatened musk deer species. All musk deer populations are decreasing, primarily because of poaching for musk pods, but also as a result of habitat degradation and hunting for meat consumption. Reliable information on each species’ distribution will help guide enforcement agencies such as local forest departments and management authorities to formulate appropriate strategies for in situ and ex situ conservation.
In addition, a range-wide population assessment will help identify poaching hotspots and combat wildlife trafficking. Therefore, we recommend extensive field sampling, including the recording of ecological data and photographic evidence, to clarify the distribution limits of the various musk deer species in the Indian subcontinent. All musk deer species should be treated as distinct evolutionary significant units, requiring long-term monitoring and evidence-based management. Reliable information about the distribution of cryptic species such as musk deer is crucial for implementing effective laws to protect and manage them.
We provide the first evidence of M. cupreus from Kedarnath Wildlife Sanctuary and Nanda Devi Biosphere Reserve in Uttarakhand, India, based on genetic analysis of the mtDNA control region. Our findings provide new information on the species' geographical distribution and will aid in formulating effective conservation strategies for this Endangered species. We recommend revising the distribution range of M. cupreus in the IUCN Red List record for this species, to support evidence-based management of musk deer in the region. Future research should include a comprehensive ecological and molecular assessment, with high throughput sequencing and microsatellite markers, and molecular tracking of confiscated items in the wildlife trade. A collaborative study across all range countries of musk deer species is vital for a comprehensive population and distribution assessment of these threatened species.
Acknowledgements
This work is part of research initiated under the Wildlife Forensic and Conservation Genetics Cell, Wildlife Institute of India. We thank Dhananjai Mohan (Director of the Wildlife Institute of India), Y.V. Jhala (Dean), and G.S. Rawat (former Dean and Director) for their support; the State Forest Departments of Jammu and Kashmir and Uttarakhand for forwarding biological samples; Aishwarya Ramachandran for map preparation; and A. Madhanraj and the Wildlife Forensic and Conservation Genetics Cell team for their support.
Author contributions
Conceptualization of study: SKG, AK, BS; data analysis: AK, BS, SS, KBG; writing: all authors; revision: AK, SKG.
Conflicts of interest
None.
Ethical standards
This research abided by the Oryx guidelines on ethical standards. All tissue samples used in this study were collected and forwarded by the State Forest Department of Uttarakhand and Jammu and Kashmir to the Wildlife Institute of India. All samples were collected from dead animals; approval from an institutional animal ethics committee was thus not required.