INTRODUCTION
Staphylococcus aureus is a leading cause of various purulent infections in humans and animals and is one of the most significant pathogens causing intramammary infections in dairy cattle worldwide. Some strains are able to produce enterotoxins (SE), which belong to the family of staphylococcal and streptococcal pyrotoxins [Reference Balaban and Rasooly1]. These toxins can cause food poisoning, toxic shock and also allergic and autoimmune reactions [Reference Balaban and Rasooly1–Reference Schlievert3]. The classical enterotoxins SEA-SEE have been well known as agents of food poisoning for several years. The emetic potential of S. aureus expressing the enterotoxin genes seg to sej is also evident, but their relevance in food intoxication remains unclear [Reference Balaban and Rasooly1]. To date, only SEH has been linked to an outbreak due to consumption of Brazilian cheese [Reference Pereira4]. In recent years a further 11 genes have been identified: sek, sel, sem, sen, seo, sep, seq, ser, seu, seu2 and sev [Reference Thomas5, Reference Thomas6]. These toxins lack the typical emetic effect although they appear to be enterotoxin-like superantigens. The toxic-shock syndrome toxin (TSST-1), a distantly related protein and the first toxin found to be involved in the toxic-shock syndrome of humans and animals, was also shown to be non-emetic [Reference Schlievert3].
The monitoring of strains in human and animal reservoirs provides important epidemiological information and several molecular methods have enabled the analysis of marker genes and facilitated the tracing of strains from various sites. Genotyping methods such as ribotyping, restriction fragment length polymorphism (RFLP), pulsed-field gel electrophoresis (PFGE), amplified fragment length polymorphism (AFLP), and multilocus sequence typing (MLST) have been all been applied to phylogenetic and epidemiological studies [Reference Thomas6] and are able to discriminate, to a greater or lesser degree, between unrelated strains [Reference Melles7].
Data from Austria regarding toxin production and antibiotic resistance of human and non-human S. aureus strains have been published separately in previous studies but the results of these investigations cannot be compared directly as different methods were used and they focused on different traits of S. aureus [Reference Winter and Baumgartner8–Reference Daxboeck10]. We set out to characterize Austrian S. aureus strains collected from veterinary, dairy and human origin in parallel, with respect to important infection-related traits such as presence of toxin genes, toxin expression, and antimicrobial resistance. Our aim was to gain comparable information about the ecological diversity of staphylococcal populations circulating in human and non-human reservoirs.
MATERIALS AND METHODS
S. aureus strains
Ninety-one veterinary strains, 247 dairy strains, and 48 human strains were included in the study. The veterinary strains originated from cases of bovine mastitis in 2001 and 2002 in Upper Austria that were investigated by the Agency for Health and Food Safety (AGES), Vienna. The dairy strains were isolated from cheese-making milk and raw milk cheeses in the period 1991–1998 by the Federal Institute for Alpine Dairying, Rotholz, and the human strains were from wound infections in 2002 at the Vienna General Hospital. All strains were from separate clinical cases or products.
The presence of clumping factor, egg-yolk reaction, and production of thermonuclease and staphylococcal enterotoxins (SE) A–E was determined for all strains but subsets of 91 veterinary, 86 dairy, and 48 human strains were tested for antimicrobial susceptibility and the presence of se genes and tst by polymerase chain reaction (PCR) analysis. These strains were also subjected to genotyping by AFLP.
Clumping factor, egg-yolk reaction, and thermonuclease
The presence of clumping factor was determined using the staphylase test (Oxoid, Basingstoke, UK). The egg-yolk reaction was tested for on Baird–Parker agar plates (Oxoid) which were incubated at 37°C and inspected after 48 h. The production of thermonuclease was detected according to OENORM-DIN 10197 on Toluidine Blue O agar [11].
Antimicrobial susceptibility testing
Susceptibility testing was performed using the agar disc diffusion method on Mueller–Hinton agar (Oxoid) as recommended by the CLSI [12, 13]. For an easier visual interpretation the DIN 58940 protocol was used for oxacillin testing [14]. The following antimicrobial agents and doses were used: amikacin (30 μg), amoxicillin/clavulanic acid (20/10 μg), cefoperazone (30 μg), clindamycin (2 μg), erythromycin (15 μg), enrofloxacin (5 μg), fusidic acid (10 μg), gentamicin (10 μg), ofloxacin (5 μg), oxacillin (5 μg), penicillin G (10 U), vancomycin (30 μg), all obtained from Oxoid, cefquinom (10 μg) from Intervet (Boxmeer, The Netherlands), and marbofloxacin (5 μg) from Vétoquinol (Lure, France). Antimicrobial zone diameters were interpreted according to CLSI, except for cefoperazone, cefquinom, and enrofloxacin in which cases the interpretation criteria of Peracef® (Pfizer, Zürich, Switzerland), Cobactan® (Intervet, Unterschleißheim, Germany) and Baytril® (Bayer, Leverkusen, Germany) were used, respectively.
Immunological detection of enterotoxin production
All strains were tested for the production of enterotoxins A, B, C, D, and E in brain heart infusion broth (BHI; Merck, Darmstadt, Germany) using the Staphylococcal enterotoxin visual immunoassay (VIA™; Tecra International Pty Ltd, French Forest, Australia).
DNA isolation
Genomic DNA was isolated from 1-ml volumes of an overnight culture at 37°C in BHI using the Qiagen® Genomic DNA kit (Qiagen Inc., Valencia, CA, USA) in accordance with the manufacturer's instructions and eluted with ddH2O. The DNA concentration was determined in a Hoefer DyNA Quant200 apparatus (Pharmacia Biotech, San Francisco, CA, USA) and adjusted with ddH2O to 1 ng/μl (PCR) or 5 ng/μl (AFLP).
Detection of enterotoxin genes and tst by PCR
Strains were tested for the presence of sea, seb, sec, sed, see, seg, seh, sei, sej, and tst by PCR as published previously, with modifications [Reference Mehrotra, Wang and Johnson15–Reference Monday and Bohach17]. Primers were obtained from MWG Biotech (Ebersberg, Germany). Primers were combined to obtain two different sets of multiplex PCRs: one set contained primers for sea, seb, sec, sed, and see (PCR 1) and the other one for seg, seh, sei, and sej (PCR 2). PCR for the detection of tst was performed separately (PCR 3). Primers and PCR conditions are shown in Tables 1 and 2. PCR amplification was carried out in a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA). Amplicons were visualized in UV light after electrophoresis in 1·5% agarose gels.
Table 1. Sequences and modifications of oligonucleotides used for PCR and AFLP
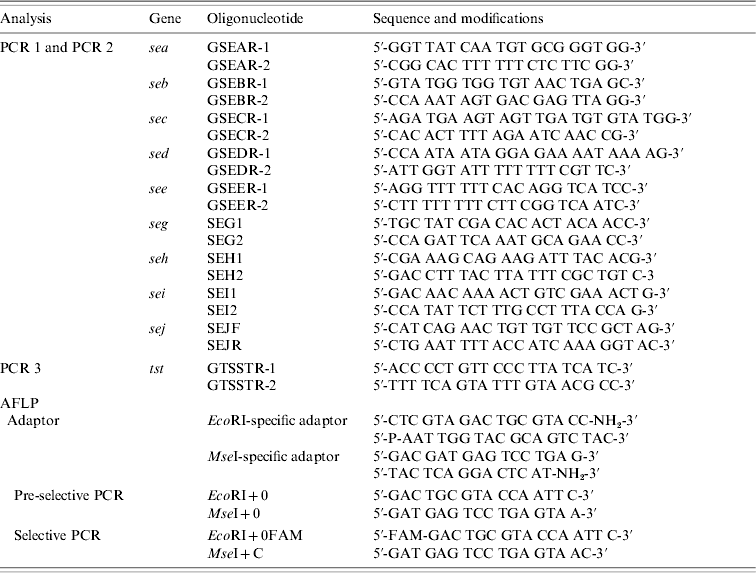
AFLP, Amplified fragment length polymorphism; PCR, polymerase chain reaction.
Table 2. PCR conditions used for the amplification of tst and see genes and for pre-selective and selective PCR (AFLP analysis)
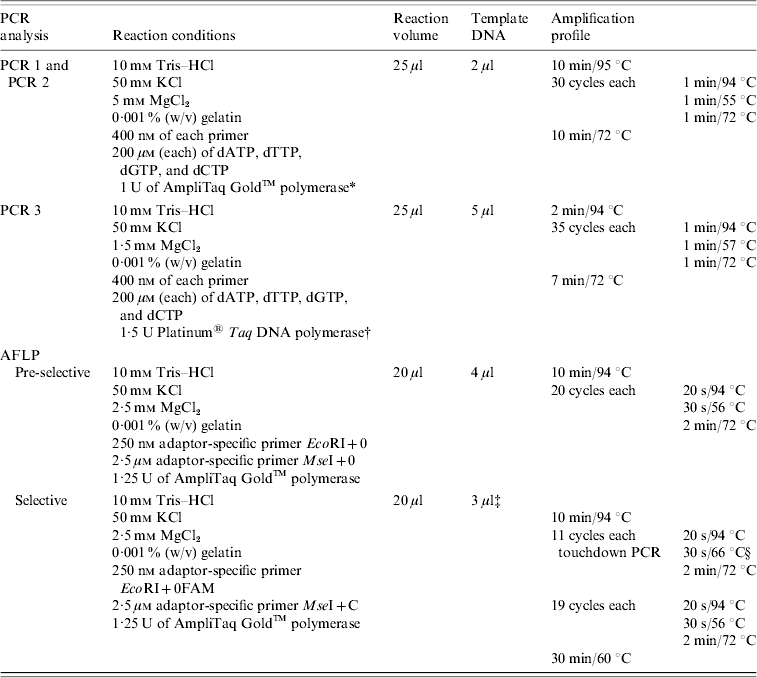
AFLP, Amplified fragment length polymorphism; PCR, polymerase chain reaction.
* Applied Biosystems, Foster City, CA, USA.
† Invitrogen, Lofer, Austria.
‡ Amplicons from pre-selective PCR diluted 1:20 with 0·1×TE buffer.
§ Decreasing 1°C every cycle until 56°C was achieved.
AFLP subtyping of S. aureus isolates
A total of 25 ng DNA was digested and ligated for 2 h at 37°C in a volume of 11 μl containing 1 μl enzyme master mix [50 mm NaCl, 50 mm Tris–HCl (pH 7·8), 10 mm MgCl2, 10 mm DTT, 1 mm ATP, 25 μg/ml BSA, 1 U MseI, 5 U EcoRI, and 1 U T4 DNA ligase (New England Biolabs, Beverly, MA, USA)], 1 μl 10×T4 DNA ligase buffer [500 mm Tris–HCl (pH 7·8), 100 mm MgCl2, 100 mm DTT, 10 mm ATP, 250 μg/ml BSA], 1 μl 0·5 m NaCl, 0·5 μl BSA solution (1 mg/ml), 1 μl EcoRI restriction site-specific adaptor (2 μm) and 1 μl MseI restriction site-specific adaptor (20 μm), 0·5 μl ddH2O, and 5 μl DNA solution (5 ng/μl). The adaptors were modified to allow exponential amplification of heterosite fragments only [Reference Lan and Reeves18]. Then 189 μl 0·1×TE buffer was added to the restriction ligation mixture.
Primers used for pre-selective and selective PCR and PCR conditions are listed in Table 1. Pre-selective and selective PCR was performed in a GeneAmp PCR System 9700 (Applied Biosystems) with the ramp rate set to 90%.
Amplified DNA fragments were separated by capillary electrophoresis on an ABI-310 Genetic Analyser (Applied Biosystems) with POP4 polymer and GeneScan™ 500 ROX size standard (Applied Biosystems) as the internal standard for each sample. Cluster analysis was performed with the Fingerprinting II software (Bio-Rad, Hercules, CA, USA) using the following settings: band classes from 120 to 400 bp, 0·5% optimization, 0·3% position tolerance, Dice similarity coefficient. For dendrogram construction the UPGMA algorithm was used. In addition, a principal components analysis (PCA) based on the binary data of the AFLP analysis was performed using the xlstat program (Addinsoft, New York, NY, USA).
Statistical analysis
Differences in the representation of strains with distinctive traits from different origins and within different AFLP clusters were calculated using Fisher's exact test or χ2 test, as appropriate (SPSS 13.0 statistical software package; SPSS Inc., Chicago, IL, USA). Heterogeneity of the AFLP profiles of different origins was calculated as published previously using Simpson's index of diversity [Reference Hunter and Gaston19].
RESULTS
Clumping factor, egg-yolk reaction, and thermonuclease production
All veterinary strains, 92·3% dairy strains, and 97·9% human strains were clumping factor-positive. Regarding the egg-yolk reaction, 53·8% veterinary strains, 45·3% dairy strains, and 85·4% human strains were positive (veterinary/human and dairy/human, P<0·0005; veterinary/dairy, P=0·165). As expected, almost all strains produced thermonuclease and only two veterinary and one human strain were negative.
Antimicrobial susceptibility testing
All 225 S. aureus strains tested were susceptible to amikacin, amoxicillin/clavulanic acid, cefoperazone, cefquinome, oxacillin, and vancomycin. A total of 136 (60·4%) of the strains were susceptible to all antimicrobial agents tested, including 84·6% of veterinary, 57·0% of dairy, and 20·8% of human strains. These differences were significant (P<0·0005). Eleven (12·1%) of the veterinary strains tested were resistant to penicillin G and two strains (2·2%) to fusidic acid. One strain was resistant to both of these antibiotics tested. Penicillin resistance was highest among dairy strains with 35 (40·6%) expressing resistance to this agent; one strain was resistant to erythromycin and another was resistant to penicillin G and fusidic acid. Resistance was most prevalent in human strains; 32 (66·7%) were resistant to penicillin G, one strain was resistant to erythromycin and four (8·3%) strains to both erythromycin and penicillin G. Only one human strain was resistant to more than two different antibiotics: clindamycin, enrofloxacin, erythromycin, gentamicin, marbofloxacin, ofloxacin, and penicillin G.
Production of enterotoxin A, B, C, D, E, and presence of se genes and tst
ELISA-based testing revealed that of 386 strains tested, 60 (15·5%) produced enterotoxins (Table 3): 9·9% veterinary, 12·6% dairy, and 41·7% of human strains. The latter were statistically significant (P<0·0005), unlike the difference between veterinary and dairy strains (P=0·502). Veterinary and human strains were found to produce only one kind of enterotoxin, whereas 26 (83·9%) of SE-positive dairy isolates produced one, four (12·9%) produced two (SEA/SED), and one produced three different enterotoxins (SEA/SEB/SEC). None of the strains produced SEE, and SEC was the most frequently detected.
Table 3. SE (enterotoxin) phenotypes of veterinary, dairy, and human S. aureus strains
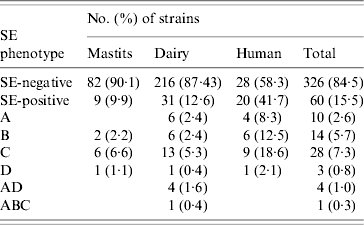
Results of PCR for enterotoxin genes of a subset of 225 S. aureus strains are shown in Table 4. Overall, 115 (51·1%) strains tested positive for one or more se gene: 40·7% veterinary, 47·7% dairy, and 77·1% of human strains. These differences were also significant (veterinary/human, P<0·0005; dairy/human, P=0·001), except for the difference between veterinary and dairy strains (P=0·596). Seven distinct se genotypes were observed in veterinary strains, 16 in dairy strains and 13 in human strains. Twenty-six (22·6%) strains harboured only one kind of se gene, the remaining 89 (77·4%) isolates possessed more than one se gene. None of the strains harboured see. The majority of strains possessed the seg gene (86 strains or 74·8% of se-positive strains), to a large extent (96·5% of seg-positive strains) in combination with other se genes. Sec was encountered most often in strains harbouring only a single se gene. The most common se genotype was seg/sei (48 strains or 41·7% of se-positive strains) followed by sec/seg/sei (16 strains or 13·9% of se-positive strains). Strains with more than three se genes were scarce: two dairy strains of se genotype sec/seg/sei/sej and one dairy strain of se genotype sea/sed/seg/sei/sej. Corresponding genes to the ELISA-based phenotypes could be detected in all cases but one dairy strain that produced SEA tested negative for the sea gene. Conversely three veterinary strains which were positive for sec did not have the corresponding phenotype, as did one sed-positive dairy strain, one sec- and one seb-positive human strain.
Table 4. Enterotoxin genotypes of veterinary, dairy, and human S. aureus strains
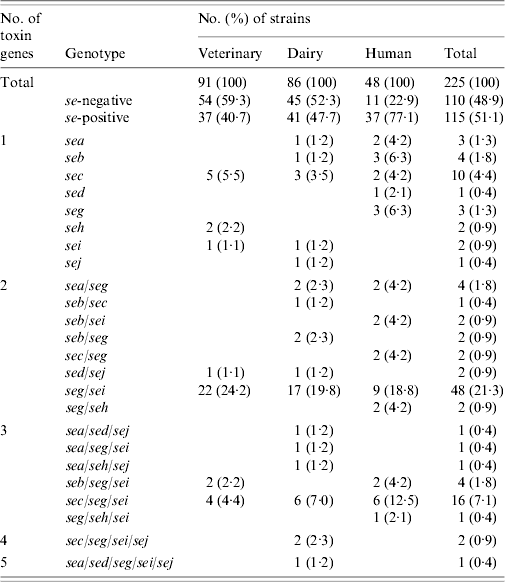
Tst was present in a total of 122 (54·2%) strains: 35 (38·5%) veterinary, 45 (52·3%) dairy, and 42 (87·5%) human strains. The difference between veterinary and dairy strains was not significant (P=0·129). However, significantly more tst-positive strains were se-positive than tst-negative strains: 71 (58·2%) of 122 tst-positive strains vs. 44 (42·7%) of 103 tst-negative strains (P=0·021). The frequency was significantly different for seb (P=0·041), sec (P=0·001), seg/sei (P=0·014), and sec/seg/sei (P=0·035) genotypes.
AFLP-based cluster analysis
Combined cluster analysis of 91 veterinary, 86 dairy, and 48 human strains revealed 17 clusters (A–Q) at a homology cut-off of 80% (Fig. 1). The largest cluster (D) contained 47·1% of all strains, followed by cluster I (13·8%). and cluster G (8%). The other clusters comprised 2–10 strains. Sixteen strains (7·1%) were not included in any of these clusters at this cut-off.

Fig. 1. Amplified fragment length polymorphism (AFLP)-based cluster analysis of veterinary, dairy and human strains. Origin and antimicrobial susceptibility of the strains are indicated by the colour of the respective boxes. Red, veterinary; green, dairy; violet, human; yellow, resistant; blue, susceptible.
Nine clusters with over- or under-representation of strains with specific characteristics were identified (Table 5). However, five of these were represented by only 2–8 strains. The largest cluster (D) contained veterinary, dairy and human strains. Under-representation of se-positive strains was mainly due to lower frequencies of this trait in dairy and veterinary strains, whereas under-representation of penicillin-susceptible strains was mainly due to lower frequencies in human and to a lesser extent in dairy strains of this cluster. Interestingly, 7/12 penicillin-resistant veterinary strains were found in this cluster. The second largest cluster (I) comprised almost exclusively dairy and human strains. Over-representation of se- and sea-positive strains was due to a higher frequency of these traits in both, dairy and human strains assembled in this cluster. By contrast, over-representation of tst-positive and antimicrobial-resistant strains was due to the lack of veterinary strains in this cluster. In addition, strains in cluster I had a higher frequency of antimicrobial resistance than generally observed for both origins. Under-representation of sei- and seg/sei-positive strains was due to the lower frequency of these genes in the strains of cluster I and to the lack of veterinary strains. Thus this cluster represented an assembly of dairy and human strains with specific traits.
Table 5. Statistically significant differences in the distribution of strains with specific characteristics between AFLP clusters
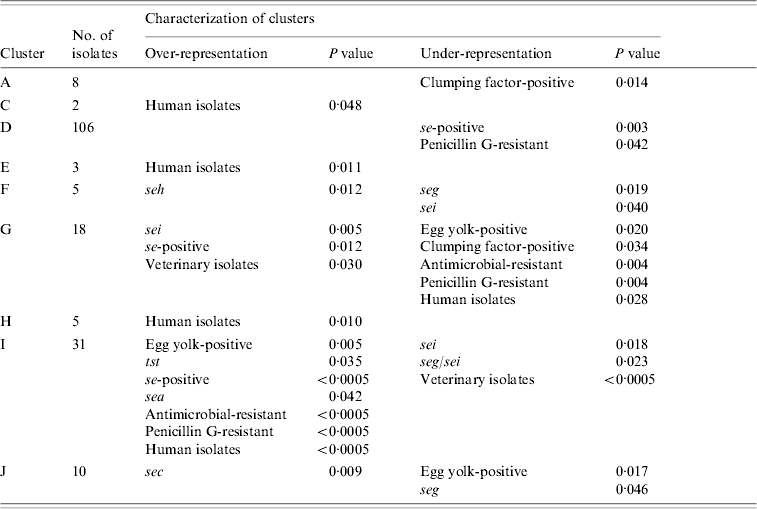
AFLP, Amplified fragment length polymorphism.
Over- and under-representation of specific se genes was calculated with respect to the number of se-positive strains of each origin.
The third major cluster (G) was almost exclusively made up of veterinary and dairy strains with more se- and sei-positive strains than observed for all strains of these origins. Under-representation of strains with antimicrobial resistance can be explained by a lower resistance rate in the dairy and veterinary strains of this cluster as well as the complete lack of human strains. Overall these findings suggest that the strains of cluster G might have grouped according to origin as well as specific traits. Over-representation of sec in cluster J comprising veterinary, dairy, and human strains was due to a higher frequency of sec in veterinary and dairy strains. Similarly, under-representation of seg could be attributed to a lower frequency of this gene in veterinary and dairy strains assembled in this cluster.
Representation of the AFLP results in a diagram defined by the first two principal components from a correlation matrix based on the binary AFLP data revealed close correlation between all strains (Fig. 2). Two separate groups were observed: a large group containing almost all strains and a small group containing 16 strains from veterinary and dairy origin belonging to clusters G, I, L, N, O, Q, or to no cluster. These strains did not have any specific phenotypic or genotypic traits in common. Human strains were not homogeneously distributed throughout the large group.The degree of genetic heterogeneity was similar for veterinary (0·701), dairy (0·733), and human strains (0·712).

Fig. 2. Distribution of (a) veterinary, dairy, and human strains, (b) antimicrobial resistant and susceptible strains, and (c) strains belonging to different UPGMA clusters in a diagram defined by the first two principal components from a correlation matrix based on the binary amplified fragment length polymorphism (AFLP) data.
DISCUSSION
The aim of the present study was to compare phenotypic and genotypic traits of S. aureus strains collected from veterinary, dairy, and human origin in order to gain information about the differences (or similarities) between strains from these sources and also between our data and those from other countries.
Clumping factor, egg-yolk reaction, and thermonuclease production
The percentage of clumping factor-positive (100%) veterinary strains was in accordance with recently published data [Reference Zschöck20, Reference Cenci-Goga21]. About two-fold more egg-yolk-positive veterinary strains (53·8%) were observed than reported in the literature [Reference Hata22, Reference Stephan, Dura and Untermann23] but the frequency of clumping factor (94·7% ) and egg-yolk reaction (43·5%) compares with other studies [Reference Müller24]. However, Becker et al. [Reference Becker, Gang-Stiller and Terplan25] reported only 44·2% of strains from quarter milk samples were positive for clumping factor. The high percentage of clumping factor-positive human strains is in agreement with other studies but here fewer egg-yolk-positive strains (85·4%) than reported by Umeki et al. [Reference Umeki26] (98·5%) were detected. The overall low percentage of egg-yolk-positive strains implies that this characteristic might not be reliable for identification of the origin of S. aureus strains. As expected, almost all strains produced thermonuclease [Reference Kloos, Schleifer, Sneath, Mair, Sharpe and Holt27].
Antibiotic resistance of S. aureus
The antimicrobial resistance rates in veterinary strains, especially penicillin resistance (12·1%), are comparable with data from Switzerland [Reference Stephan, Dura and Untermann23, Reference Roesch28]. However, resistance rates were lower than described for Austria in 1996–1998 (20·9–28·4%) but similar to the results reported for 1987–1988 [Reference Winter and Baumgartner8, Reference Aarestrup and Jensen29]. Fitzgerald et al. [Reference Fitzgerald30] recovered more penicillin-resistant S. aureus strains from bovine intramammary infections in Ireland than reported in the present study. It is well documented that the level of penicillin resistance differs between European countries, perhaps reflecting different antimicrobial prescribing policies in each country [Reference Vintov31]. The rates of penicillin, erythromycin and marbofloxacin resistance among the dairy strains accord with those from Hungary [Reference Kaszanyitzky32] while for human strains the results are in agreement with those reported in 2005 and 2006 for Austria and Germany [33, Reference Layer34], but rates were lower than reported from 1996 to 2002 for Austria [Reference Daxboeck10]. On the other hand, a higher percentage of penicillin- and erythromycin-resistant strains was observed for 13 European countries in 2000, with only 2% of strains being multiresistant, which is supported by the present study [Reference Kitzrow35]. Differences between the data from earlier Austrian studies and from other countries might reflect changing trends in antibiotic resistance but they might also result from different sampling strategies and protocols to detect antibiotic resistance.
Production of enterotoxin A, B, C, D, E, and presence of se genes and tst
The frequency of enterotoxin-producing veterinary strains (9·9%) is close to data published for California in 2003 [Reference Cenci-Goga21], and for enterotoxigenic dairy strains (12·6%), this accords with results for France in 1997 [Reference Rosec36]. SEC was most frequently produced by veterinary and dairy strains and this is supported by other studies [Reference Jorgensen37, Reference Stephan38]. Enterotoxin-producing human strains (41·7%) were considerably more frequent than veterinary (9·9%) and dairy strains (12·6%) and similar to recently reported rates [Reference Boerema, Clemens and Brightwell39]. In other studies the prevalence of SEs and also the frequency of each SE were highly variable [Reference Cenci-Goga21, Reference Stephan38] and this might be explained by variation in assays and culture conditions used to detect enterotoxins. The absence of SEE and the see gene accords with the results of others [Reference Boerema, Clemens and Brightwell39, Reference Morandi40].
PCR assays showed good correlation with the expression of enterotoxin proteins detected by ELISA but PCR identified a higher number of enterotoxin-positive strains. The finding that in one strain SEA could not be detected by PCR might be explained by sequence variation in the sea gene or a novel toxin cross-reacting in the ELISA [Reference McLauchlin16, Reference Jorgensen37]. The most common combination within all strains was seg/sei followed by sec/seg/sei. Similar results were found in other European countries [Reference Becker41, Reference Stephan, Buehler and Lutz42]. In animal-associated strains sea, seb, see, and seh were absent or rarely present as commonly observed [Reference Cenci-Goga21, Reference Fitzgerald30]. A small percentage of human strains harboured the classical enterotoxin genes sea, seb or sec, but seg and sei were found in 50% and 41·7% of human strains, respectively. The function of the recently described toxin genes as pathogenic agents remains unclear but S. aureus strains involved in food-poisoning outbreaks lacking classical enterotoxins but expressing seg, sei, and sej have been described [Reference McLauchlin16].
The proposed coexistence of toxin genes sed and sej on a common plasmid was not supported by our data [Reference Zhang, Iandolo and Stewart43]. Similar results were found by others [Reference Morandi40, Reference Cremonesi44]. Coexistence of seg and sei on the egc enterotoxin cluster in S. aureus is also controversial [Reference Layer34, Reference Jorgensen37, Reference Morandi40, Reference Cremonesi44]. In the present study seg was detected in 84·9% of all cases together with sei. However, sequence variation might also account for these results.
The frequency of tst among veterinary strains (38·5%) was lower than reported for Switzerland (64·7%) [Reference Stephan38] but the percentage of tst-positive dairy strains (52·3%) was slightly higher than found recently in Norway [Reference Jorgensen37]. Contrary to studies performed in other countries which have reported 6·5–40% tst-positive human strains, we found most (87·5%) human strains were tst positive [Reference Layer34, Reference Leuween45, Reference Peacock46]. An alternative PCR method was performed on selected strains and confirmed the results, thus non-specific PCR amplification could be ruled out (data not shown) [Reference Monday and Bohach17]. In our study 20·5% of all tst-positive strains were positive for sec and only 2·6% of sec-positive strains carried no gene for tst. Significant correlation between SEC and TSST has been reported for bovine strains [Reference Fitzgerald30, Reference Stephan38], perhaps due to the fact that both genes are located on the bovine staphylococcal pathogenicity island (SaPlbov), a mobile genetic element [Reference Fitzgerald47]. Our investigations showed that of nine veterinary strains carrying the gene for sec, seven also carried tst. In addition, only one of 12 sec-positive dairy strains had no tst gene. The rare co-occurrence of sec and tst genes in human strains has been reported [Reference Becker41, Reference Peacock46]. This contrasts with our finding of 80% of sec-positive human strains also carrying tst.
AFLP-based cluster analysis
Clusters D, G, I, and J comprised almost 70% of all strains and contained strains with different frequencies of se genes or antimicrobial resistance than observed for all strains of each origin. In addition, two of these clusters grouped strains according to origin, since in both cases mainly two niches were represented in the cluster. Recently published AFLP analysis of S. aureus strains from human and animal origin also revealed a tendency to cluster according to origin whereas single enzyme AFLP of human, bovine, ovine, and food-related sources revealed a tendency to cluster according to se genotypes [Reference Boerema, Clemens and Brightwell39, Reference Leuween45].
In conclusion, many of the phenotypic and genotypic traits of S. aureus strains from veterinary, dairy, and human origin were similar to those reported from other European countries. However, marked differences were observed between strains from different origins for antimicrobial resistance, presence of se genes and tst gene. An outstanding finding was the high percentage of tst-positive human strains. Since S. aureus strains characterized here were collected from human wound infections only, further research should focus on comparing the percentage of tst-positive human strains from different clinical settings and from healthy individuals. Standardized methods for characterization of S. aureus strains would greatly improve the comparison of data from different ecological niches and different countries. The European Antimicrobial Resistance Surveillance System (EARSS) for human S. aureus strains was established in 1998 with the aim of obtaining comparable and reliable data for public health. Extension of this programme to the monitoring of toxins and inclusion of veterinary strains is worthy of consideration.
ACKNOWLEDGEMENTS
The authors thank the Institute of Animal Breeding and Genetics, Clinical Department of Animal Breeding and Reproduction, University of Veterinary Medicine, Vienna, Austria for kindly providing the ABI-310 Genetic Analyser.
DECLARATION OF INTEREST
None.









