In oviparous fish, the transition from utilisation of yolk nutrients to exogenous feed requires considerable morphological, physiological and behavioural changes( Reference May 1 ). Survival and growth of a developing larva is determined by the development of functional organ systems and availability and quality of feed( Reference Sifa and Mathias 2 – Reference Busch, Falk-Petersen and Peruzzi 4 ). Rotifers (mainly Brachionus sp.) followed by brine shrimp (Artemia salina) are the live prey most widely used for start-feeding of marine fish larvae in aquaculture. However, their nutritional value is suboptimal, resulting in inferior growth and performance compared with larvae fed natural zooplankton, such as in the case of Atlantic cod (Gadus morhua) larvae( Reference Imsland, Foss and Koedijk 3 , Reference Busch, Falk-Petersen and Peruzzi 4 ).
Studies on mammals and fish have shown that feed nutrient composition affects differential gene expression and activation of various metabolic pathways( Reference Tacchi, Secombes and Bickerdike 5 , Reference Penglase, Edvardsen and Furmanek 6 ), including modulation of microRNA (miRNA)( Reference Parra, Serra and Palou 7 , Reference Beckett, Yates and Veysey 8 ). miRNA are approximately twenty-two-nucleotide-long regulatory RNA present in animals and plants. They mainly pair with sites in the 3′ untranslated regions (3′ UTR) of mRNA to mediate post-transcriptional repression( Reference Bartel 9 ). The extent of sequence complementarity between miRNA and mRNA leads to control of protein production in the cell by blocking translation, degradation of the mRNA or both( Reference Djuranovic, Nahvi and Green 10 ). Many miRNA have pleiotropic effects and mediate a variety of physiological processes including differentiation, growth, development and energy metabolism( Reference Dumortier, Hinault and Van Obberghen 11 , Reference Bizuayehu and Babiak 12 ). In addition, miRNA expression is under less strict genetic control than that of mRNA( Reference Parts, Hedman and Keildson 13 , Reference Siddle, Deschamps and Tailleux 14 ), which indicates a possible regulation of miRNA by epigenetic mechanisms.
Metabolic regulation is a principal mechanism to control differentiation, proliferation and survival of cells( Reference Williams, Henao-Mejia and Harman 15 ). Metabolic regulation depends on different intrinsic and extrinsic factors, including availability of nutrients. Lack of essential nutrients is among the major reasons of lower growth rate and deformities in Atlantic cod larvae fed rotifers compared with natural zooplankton diets( Reference Hamre 16 , Reference Imsland, Foss and Koedijk 3 ). miRNA act as essential metabolic rheostats at a cellular level( Reference Henao-Mejia, Williams and Goff 17 ) and can boost mitochondrial ATP production( Reference Zhang, Zuo and Yang 18 ). Therefore, these versatile regulatory molecules likely mediate nutritional effects on gene expression. However, limited information exists on miRNA control of metabolism and involvement in nutrient utilisation in teleost fish( Reference Mennigen, Panserat and Larquier 19 – Reference Mennigen, Plagnes-Juan and Figueredo-Silva 21 ). In juvenile rainbow trout (Oncorhynchus mykiss), expression of miR-122, a liver-specific miRNA, was altered with a change in dietary macronutrient ratio, particularly carbohydrate:protein and lipid:protein ratio( Reference Mennigen, Plagnes-Juan and Figueredo-Silva 21 ). Similarly, miR-143 expression increased after the commencement of exogenous feeding in rainbow trout alevins( Reference Mennigen, Skiba-Cassy and Panserat 20 ). However, the only information on the link between miRNA regulation and the type and composition of larval feed is available from a study on rainbow trout alevins fed diets containing a high or very low protein:carbohydrate ratio, where the four studied miRNA – miR-29, miR-33, miR-107 and miR-143 – showed no differences in expression( Reference Geurden, Mennigen and Plagnes-Juan 22 ). Previous experiments and observations have shown that the type of live feed has a profound effect on development as well as lifelong growth of Atlantic cod( Reference Imsland, Foss and Koedijk 3 ).
This study provides a first insight into the nutrient-driven miRNA repertoire in fish larvae fed different types of live prey as a start-feed. Relationships between the differentially expressed miRNA and their putative target mRNA were also investigated. The results of this study provide some useful information on the role of miRNA in a metabolic network during fish larval development.
Methods
Fish, experimental set-up and sampling
The experiment was conducted at Austevoll Research Station of the Institute of Marine Research, Storebø, Norway (IMR-Austevoll). Care and handling of fish used in this study were in accordance with the Norwegian Regulation on Animal Experimentation (The Norwegian Animal Protection Act no. 73 of 20 December 1974, Section 20–22, amended 19 June 2009). The specific experiment described in this study was approved by the local representative of the Norwegian Research Animal Committee (Forsøksdyrutvalget, www.fdu.no) with the acceptance reference FOTS id 5448.
A detailed description of experimental set-up, including live feed production and larval rearing, is described in the study by Karlsen et al. ( Reference Karlsen, van der Meeren and Rønnestad 23 ) and van der Meeren et al. ( Reference van der Meeren, Karlsen and Liebig 24 ). Eggs were obtained from forty-five females and twenty-five males of Atlantic cod kept under natural photoperiod in a 7-m diameter indoor fibre-glass tank (average temperature: 7·9 (SD 0·3)°C and salinity: 34·7(SD 0·2) g/l). The brood fish were 3 years old and in the range between 2 and 3 kg, originating from broodstock maintained at IMR-Austevoll. They were fed Skretting Vitalis diet (Skretting) three times a week. Fish were spawning spontaneously every night during March and April 2012. Upon spawning, fertilised floating eggs were concentrated overnight in outlet collectors, disinfected with glutaraldehyde and incubated in six 70 litre flow-through egg incubators with gentle air bubbling around a centrally mounted outlet screen, at 6°C under continuous 24-h roof light of 33 μW/cm2 intensity, until beyond hatching.
At 4-d post-hatch (dph), larvae were transferred from incubators to six black 400 litre start-feeding tanks. An equal number of larvae from each incubator (approximately 12 500 larvae/incubator; 50 000 in total) were stocked in each start-feeding tank (approximately 125 larvae/l). Before transfer, the start-feeding tanks were supplied with algal paste to produce turbidity (green water), beneficial for larval feeding( Reference van der Meeren, Mangor-Jensen and Pickova 25 ). Rotifers or zooplankton (various stages of copepods) were provided as a live feed separately to three tanks each, before larval transfer.
The tanks were divided into two treatment groups: the aquaculture feed group receiving the most common marine hatchery feed (rotifers, substituted later on with Artemia) and the reference group, which was fed zooplankton harvested from a seawater pond, predominately nauplii and copepodids of Acartia longiremis and Centropages hamatus ( Reference van der Meeren, Karlsen and Liebig 24 ). In the first treatment, rotifers were enriched with LARVIVA Multigain (BioMar) and Sel-Plex® (Alltech) used as feed from 4 to 32 dph, with a gradual shift to Multigain-enriched Artemia from 32 to 35 dph, continuing with Artemia until 63 dph. Gradual switch to the formulated diet AgloNorse (Tromsø Fiskeindustri AS) took place from 58 to 63 dph. In the second treatment, larvae were fed zooplankton obtained from the seawater pond ‘Svartatjern’ (Austevoll, Norway). This comprised nauplii from 4 to 30 dph, copepodids from 18 to 44 dph and gradual switch to AgloNorse from 37 dph. The amount of feed was adjusted daily by visual observation of the remaining prey in the tanks after feeding and evaluation of stomach fullness. Larvae were reared under 16 h:8 h photoperiod and light intensity of 300–500 μW/cm2. The temperature was increased gradually from 8 to 11·6°C, from day 4 to day 11 post-hatch, respectively, more than 10 d before any observed differences in growth rates. Initial water flow was 1 l/min; it increased to 6 l/min at switch to the formulated diet and to 10 l/min at experiment termination( Reference Karlsen, van der Meeren and Rønnestad 23 ).
Sampling dates in the two treatment groups were adjusted to larval size. Samples were collected when fish reached an average standard length of 4·5, 5·2, 7·0, 9·3, 13·9 and 25·1 mm, which corresponded to 4, 11, 22, 29, 37 and 53 dph in the reference feed group and to 4, 11, 22, 31, 54 and 71 dph in the aquaculture feed group, respectively( Reference Karlsen, van der Meeren and Rønnestad 23 ). These six stages are named as stages 0–5 throughout this paper. Whole larvae samples were snap-frozen in liquid N2 and stored at −80°C until RNA extraction.
RNA extraction and sequencing
Total RNA was extracted using Trizol (Invitrogen) from at least six individuals from each rearing tank. Similarly, RNA was extracted from the live feeds (natural zooplankton, rotifers and Artemia nauplii). Quality of RNA was examined using a BioAnalyzer (Agilent Technologies). Samples with a 28S:18S RNA ratio ≥1·8 and RNA integrity number ≥8 were used for sequencing library preparation. Equal concentration of RNA was pooled from the replicate tanks. With the exception of stage 2 and A. nauplii samples, next-generation sequencing libraries were prepared following SOLiD Whole Transcriptome Analysis Kit protocol (Life Technologies) and sequenced using 5500xl SOLiD sequencer (Life Technologies) at the University of Nordland, Norway. Libraries from stage 2 and A. nauplii samples were prepared using NEXTflex Small RNA-Seq Kit v2 (Bioo Scientific) and sequenced using NextSeq 500 (Illumina) at the University of Oregon, USA. The use of two sequencing technologies did not affect the experimental set-up, because no temporal effects were investigated and all comparisons were performed for each stage separately. mRNA sequencing and data analysis were performed as described by Penglase et al.( Reference Penglase, Edvardsen and Furmanek 6 ).
Characterisation of microRNA and differential expression analysis
Adapter sequences and low-quality bases were trimmed using cutadapt( Reference Martin 26 ). Sequences with high-quality score (≥20) at least in the first half of the sequence were mapped to the Atlantic cod genome, gadMor1( Reference Flicek, Amode and Barrell 27 ), using Bowtie2( Reference Langmead, Trapnell and Pop 28 ) with default setting. Conserved miRNA were identified using miRDeep2( Reference Friedländer, Mackowiak and Li 29 ) by similarity searching against fish miRNA from miRBase 21( Reference Kozomara and Griffiths-Jones 30 ) and Atlantic cod miRNA( Reference Bizuayehu, Johansen and Puvanendran 31 ), whereas the search for conserved miRNA in rotifers, zooplankton and Artemia samples was performed against arthropods miRNA from miRBase 21( Reference Kozomara and Griffiths-Jones 30 ). The minimum threshold for miRNA count was set at 10 reads per million (rpm) at least in one of the samples. As RNA was extracted from the whole larvae, we also mapped small RNA sequences of Atlantic cod larvae to miRBase arthropods miRNA to check whether the miRNA that were expressed in the larvae were the same as those in the live feeds.
Differential expression of miRNA between the groups at six stages were assessed separately using NOISeq( Reference Tarazona, García-Alcalde and Dopazo 32 ). We used parameters as previously described( Reference Bizuayehu, Johansen and Puvanendran 31 ) with some differences. In this study, we considered differential expression between the groups, q>0·95 with threshold of 10 000 rpm counts difference and 0·8<q<0·95 with threshold of 15 000 rpm counts difference. We used these stringent criteria to avoid false positives. This setting is reasonable, as NOISeq has a better correct calling for over-represented than under-represented genes( Reference Zheng and Moriyama 33 ).
Real-time RT-PCR
To test the sequencing results, we performed real-time RT-PCR for the chosen miRNA: miR-19, miR-25, miR-203 and miR-204. Totally, six samples from each treatment (two pools from each tank) were used for real-time RT-PCR. Pooling was carried out by taking four random individuals for each pooled sample. Complementary DNA (cDNA) was synthesised using miRCURY LNA universal cDNA synthesis kit II (Exiqon). The reaction was performed on LightCycler 480 (Roche Applied Science) with SYBR® Green chemistry using white ninety-six-well plates with thermal cycle conditions of 95°C for 10 min, followed by forty-five cycles of 95°C for 10 s and 60°C for 1 min. Cq values were collected using second derivative method. We used 5S ribosomal RNA to normalise the expression levels of miR-19, miR-25, miR-203 and miR-204. The Shapiro–Wilk normality test was performed, and data not following normal distribution were log-transformed. The data fitting normality were analysed using Student’s t test, whereas the data that did not fit normality were analysed using non-parametric Mann–Whitney U test.
MicroRNA target prediction
To predict miRNA targets, 3′ UTR of the Atlantic cod mRNA sequences were downloaded from NCBI database. We also obtained 3′ UTR sequences from our RNA-Seq data (the Sequence Read Archive at NCBI, accession ID SRP056073). For this, we performed BLASTX search to peptide sequences of Atlantic cod Ensembl 79( Reference Flicek, Amode and Barrell 27 ) with e-value 10−4. We filtered sequences having similarity >75 % and considered only alignments covering stop codons. Next, we extracted sequences after the stop codon and annotated them as 3′ UTR. Potential miRNA-binding sites were identified using miRanda( Reference John, Enright and Aravin 34 ) with strict seed matching, score ≥150, and free energy threshold ≤−83 kJ/mol (≤−20 kcal/mol).
Functional annotation of microRNA through ortholog assignment of their targets
Targets of differentially expressed miRNA were annotated to Atlantic cod Ensembl 79( Reference Flicek, Amode and Barrell 27 ) gene ontology (GO) terms. Similarly, miRNA targets were annotated with ortholog Kyoto Encyclopedia of Genes and Genomes (KEGG) genes( Reference Kanehisa, Araki and Goto 35 ), using the best hit from a BLAST analysis against Danio rerio, Homo sapiens, Caenorhabditis elegans and Xenopus laevis with KEGG pathways annotation. Enrichment of terms was tested using g:Profiler( Reference Reimand, Arak and Vilo 36 ) for targets of differentially expressed miRNA at each stage. P values were adjusted for multiple testing by Benjamini–Hochberg correction. P<0·05 was considered as statistically significant.
Results
MicroRNA in Atlantic cod larvae and their live feed
We obtained 112 million small RNA sequences from eleven Atlantic cod libraries, and on average 59 % of them were mapped to Atlantic cod genome. miRNA constituted on average 26 % of the total small RNA sequences. In addition, rotifers, zooplankton and Artemia libraries yielded more than ninety-six million small RNA sequences, which consisted of 0·44, 10 and 13 % miRNA, respectively (Table 1, online Supplementary Table S1).
Table 1 Overview of small RNA sequencing dataFootnote * (Numbers and percentages)
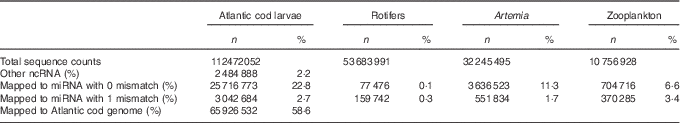
ncRNA, non-coding RNA; miRNA, microRNA.
* The total count represents clean adapter removed sequences. Sequences obtained from Atlantic cod larvae fed either aquaculture or reference live feeds (pooled five libraries each) were mapped to Atlantic cod miRNA, other ncRNA and genome; whereas sequences obtained from live feeds were mapped to arthropods miRNA (miRBase 21).
A number of conserved miRNA in both live feeds and Atlantic cod larvae were identified, but none of the arthropod-specific miRNA was considerably present in Atlantic cod larval small RNA libraries (online Supplementary Table S2). By considering 10 rpm as a minimum miRNA count, we identified 348, 165, 95 and 70 mature miRNA in Atlantic cod larvae, rotifers, Artemia and zooplankton, respectively (online Supplementary Table S2). In spite of the fact that miRNA in rotifers had the highest diversity among the live feeds, approximately 90 % of the total miRNA counts constituted of miR-87 and miR-125. The single most dominant miRNA in Artemia was bantam, whereas in the lagoon zooplankton sample the most abundant miRNA was miR-993, and the top five miRNA covered 70 % of the total miRNA counts.
Differential expression
The effects of the type of live feed on larval development and performance, nutrient composition and the redox system have been reported in separate papers( Reference Karlsen, van der Meeren and Rønnestad 23 , Reference Penglase, Edvardsen and Furmanek 6 ). Generally, fish fed zooplankton (reference group) performed significantly better in most measures – for example, they were significantly longer and heavier from 22 dph onwards and had almost twice the growth rate (4·5 v. 2·4 %/d) during this period( Reference Karlsen, van der Meeren and Rønnestad 23 ).
In the course of the experiment, eight miRNA showed significant differences in expression between the two feeding groups. At specific stages, differential expression was found for two miRNA at stage 1, six miRNA at stage 2, three miRNA at stage 3, three miRNA at stage 4 and no miRNA was differentially expressed at stage 5 (Table 2). Expressions of some miRNA (miR-19a, miR-130b, miR-181a and miR-11240) were significantly higher in the reference feed group, some others (miR-146 and miR-192) had higher expression in the aquaculture feed group and few others (miR-9 and miR-206) showed mixed, developmental stage-dependent expression in the both feeding groups.
Table 2 MicroRNA expression in Atlantic cod larvae fed aquaculture (Brachionus sp. followed by Artemia sp.) or reference (zooplankton harvested from a lagoon, mostly copepods) dietsFootnote †

Stage 0, start of experiment.
*Significantly different (for details see the ‘Methods’ section).
† Read counts are normalised and presented as reads per million.
Verification of the NOISeq analysis with real-time RT-PCR analysis indicated no significant differences between the two feeding groups, except for miR-19d, miR-25 and miR-204 expressions at stage 1 (Fig. 1). The NOISeq analysis showed similar relations between miRNA expressions without false positives in the two feeding groups.

Fig. 1 Expression of microRNA obtained by next-generation sequencing (NGS) and real-time RT-quantitative PCR (RT-qPCR) from Atlantic cod larvae fed aquaculture (Brachionus sp. followed by Artemia sp.) or reference (zooplankton harvested from a seawater pond, mostly copepods) feeds and sampled at developmental stages 0–5. For real-time RT-PCR, expression levels were normalised against 5S ribosomal RNA. Values are means (n 6), with standard deviation represented by error bars. * Significantly different between the two feeding groups within a stage: P<0·05. ![]() , Reference NGS;
, Reference NGS; ![]() , aquaculture NGS;
, aquaculture NGS; ![]() , reference RT-qPCR;
, reference RT-qPCR; ![]() , aquaculture RT-qPCR; rpm, reads per million.
, aquaculture RT-qPCR; rpm, reads per million.
Relation between differentially expressed microRNA and their potential targets
We annotated the 3′ UTR of 3698 genes, and 2770 of them had miRNA-binding sites for at least one of the Atlantic cod miRNA characterised in this study. Further analysis revealed that the eight differentially expressed miRNA could potentially target 397 mRNA (online Supplementary Table S3). Three mRNA were potential targets for three of these eight differentially expressed miRNA, and further forty-one mRNA were targets of two differentially expressed miRNA.
In all, eighteen of these 397 mRNA showed differential expression patterns across the study. They included transcriptional factors, digestive enzymes, structural proteins, kinases and transporter transcripts (online Supplementary Table S4). The reciprocal relations of expression patterns of these mRNA and differentially expressed miRNA between the two feeding groups across the study (Fig. 2, online Supplementary Fig. S1) showed two general patterns: inverted, when higher miRNA expression in one of the feeding groups was accompanied by lower mRNA target expression in the same feeding group (Table 3), and concurrent, when both miRNA and its target mRNA expression were elevated within the same feeding group (online Supplementary Fig. S1). An inverse relation was observed, for example, at stage 2, when a significant down-regulation of miR-9 and significant up-regulation of its targets insulin-like growth factor binding protein, acid-labile subunit (igfals) and chymotrypsin-like elastase family, member 2A (cela2a) was found in the aquaculture feed group as compared with the reference feed group. Similar inverse relation was found between dual-specificity phosphatase 5 (dusp5) and calmodulin-like protein 4 (calml4) and miR-181a (Fig. 2). Some other miRNA–mRNA pairs showed inverse relationships, but not significantly different between the two feeding groups in miRNA or mRNA expression at the same stage (Fig. 2). For instance, this was the case for miR-19a and collagen alpha-2(I) chain (col1a2) at stage 4, miR-206 and myb at stage 3, miR-9 and retinal G protein-coupled receptor at stage 4, as well as miR-9 and phosphorylase kinase gamma 1 (phkg1) at stage 4. Concurrent expression was observed for miR-206 and myb at stage 4 as well as miR-9 and cela2a at stage 3 (Fig. 2, online Supplementary Fig. S1).

Fig. 2 Expression of selected microRNA (miRNA) and their putative mRNA targets in Atlantic cod fed aquacultured live feed v. reference natural zooplankton live feed. At stage 5, both feeding groups were fed formulated diet. miRNA expression levels are given in reads per million (rpm), whereas mRNA reads are normalised by the total number of mapped sequences. Samples were taken at five larval stages and one juvenile stage. Values are means, with standard deviation represented by error bars. *, ** Significantly different expressions of mRNA and miRNA, respectively, between the two feeding groups within a stage (q>0·8; for details see the ‘Methods’ section). slc40a1, Solute carrier family 40 (Fe-regulated transporter), member 1; dusp5, dual-specificity phosphatase 5; calml4, calmodulin-like protein 4; cyp2k1, cytochrome P450 2K1; col1a2, collagen alpha-2(I) chain; igfals, insulin-like growth factor binding protein acid-labile subunit; c-myb, transcriptional factor myb; cryba1, crystallin, beta A1.
Table 3 Inverse expressions of microRNA (miRNA) and their putative mRNA targets in Atlantic codFootnote *
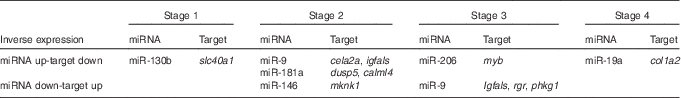
slc40a1, solute carrier family 40 (Fe-regulated transporter), member 1; cela2a, chymotrypsin-like elastase family, member 2A; igfals, insulin-like growth factor binding protein, acid-labile subunit; myb, transcriptional factor myb; col1a2, collagen alpha-2(I) chain; dusp5, dual-specificity phosphatase 5; calml4, calmodulin-like protein 4; mknk1, MAP kinase interacting serine/threonine kinase 1; rgr, retinal G protein-coupled receptor; phkg1, phosphorylase kinase, gamma 1.
* Expression in reference feed group compared with aquaculture feed group is shown. Only miRNA and their putative targets each showing significant differential expression between the two feeding groups at different stages are shown.
Enrichment for gene ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways of differentially expressed microRNA targets
In total, we identified 1291 GO terms and 146 KEGG pathways for the putative targets of miRNA identified in this study. Differentially expressed miRNA targets were assigned in 324 molecular functions, seventy-three cellular components and 949 biological processes. These targets were involved in ninety-eight KEGG pathways. One KEGG term (KEGG:00670, one carbon pool by folate) was significantly enriched (P=0·00669) at stages 3 and 4. However, non-enriched targets were involved in various metabolic and signalling pathways and transportation networks (online Supplementary Table S3).
Discussion
In intensive aquaculture of most marine fish species including Atlantic cod, rotifers are used as a first feed followed by Artemia and later on by a formulated diet. These live feeds are deficient in certain nutrients and are usually enriched before being offered to the larvae( Reference Hamre, Yúfera and Rønnestad 37 ). Nevertheless, and despite large progress in enrichment protocols, replacement of these live feeds with natural zooplankton results in better performance parameters such as survival, growth, pigmentation, stress resistance and normal development( Reference Imsland, Foss and Koedijk 3 , Reference Busch, Falk-Petersen and Peruzzi 4 , Reference Conceição, Yúfera and Makridis 38 ), which is attributed to superior nutritional composition of natural zooplankton( Reference Busch, Falk-Petersen and Peruzzi 4 , Reference Mæhre, Hamre and Elvevoll 39 ). In this study, we demonstrate that the nutritional effect of the first feed on larval and further juvenile performance of Atlantic cod may be related to a regulatory action of miRNA. However, global analysis of this regulation was limited by the resolution of the data and incomplete genomic resources. The low resolution resulted from technical limitations due to small larval size that prevented dissection of homogeneous tissues of relevance. Thus, RNA analysis was performed on pooled samples of larvae, which compromised the precision of transcriptome information. Lack of complete sequences of Atlantic cod 3′ UTR could have also affected our enrichment analysis. The 3′ UTR sequences derived in this study represented only 17 % of the Atlantic cod genes( Reference Star, Nederbragt and Jentoft 40 ). The analysis of teleost-specific miRNA generated 155·32 targets/miRNA on average( Reference Yang, Irwin and He 41 ), whereas in this study the average number of targets for differentially expressed miRNA was 53·25 with a minimum of 12 and a maximum of 112/miRNA (online Supplementary Table S3). Aside from the incomplete set of 3′ UTR used, low number of significantly enriched pathways could result from diverse roles of differentially expressed miRNA in various biological pathways and/or functional redundancy of miRNA. The majority of miRNA acts in constellation and reduces their target level slightly, which has been shown in their noise reduction and in conferring precise protein level in the cell( Reference Schmiedel, Klemm and Zheng 42 ).
In mammals, exogenous (dietary intake) miRNA are implicated in targeting low-density lipoprotein receptor adapter protein 1 ( Reference Zhang, Hou and Chen 43 ). However, this regulation was found for miRNA originating from rice (Oryza sativa) as a diet. Plant miRNA have 2′-O-methylation at their 3′ ends, which protects them from exonucleolytic digestion( Reference Yu, Yang and Li 44 ). Although extraction of RNA was performed using the whole organisms, we have not found considerable presence of live feed-specific (exogenous) miRNA. Although such effects cannot be ruled out, our data suggest that the feed components rather trigger endogenous mechanisms of gene expression in response to nutritional stimuli.
Several environmental factors such as temperature, photoperiod or feed have long-term effects on gene expression( Reference Jonsson and Jonsson 45 ). Recently, we demonstrated a long-lasting effect of embryonic thermal history on further miRNA expression in Atlantic cod, which was likely driven through an epigenetic mechanism( Reference Bizuayehu, Johansen and Puvanendran 31 ). Although we cannot rule out the effects of heterochrony or allometry on miRNA expression, as the two feeding groups elicited size difference as early as stage 2( Reference Karlsen, van der Meeren and Rønnestad 23 ), the epigenetic modulation of miRNA expression triggered by live feed components could contribute to ultimate phenotypic differences. However, we have not found a long-lasting effect of the first feed on miRNA expression in stage 5, where both feeding groups were fed the same formulated diet.
miRNA may regulate Fe homoeostasis. In this study, we found a significantly higher expression of miR-130b in the reference feed group (mostly copepod nauplii) at stage 1 as compared with the rotifer-fed group (Table 2). Ferroportin or solute carrier family 40 (Fe-regulated transporter), member 1 is one of the predicted targets of miR-130b. It encodes a cell membrane protein that functions as an Fe exporter( Reference Donovan, Brownlie and Zhou 46 ). Dietary Fe absorbed by enterocytes can be stored in ferritin or exported by ferroportin, which is located at the basolateral surface of the enterocyte. In mammals, hepcidin, a hormone synthesised and secreted by hepatocytes, is the major regulator of Fe transport by inducing the endocytosis and proteolysis of ferroportin( Reference Ganz 47 ). In several teleosts including Atlantic cod, hepcidin was identified and characterised as an antimicrobial peptide( Reference Douglas, Gallant and Liebscher 48 , Reference Solstad, Larsen and Seppola 49 ). Although there is no defined dietary requirement for minerals, previous results suggest that rotifers contain sufficient amount of Fe, which is ten times lower than that in natural zooplankton( Reference Imsland, Foss and Koedijk 3 , Reference Mæhre, Hamre and Elvevoll 39 ). Taken together, elevated expression of miR-130b in the reference feed group can be associated with translational repression of ferroportin. Thus, miR-130b may have a role in an additional or alternative mechanism for regulation of Fe homoeostasis.
The relation between miRNA and their putative targets could be through direct repressive interaction or indirect feedback or feed-forward mechanisms( Reference Tsang, Zhu and van Oudenaarden 50 ). Although miRNA mainly suppress their targets, their roles in general gene regulatory networks need not to be simply suppressive. They could have gene regulatory network-dependent functions through miRNA–target interactions. Hence, the identified miRNA–mRNA interactions in this study could be indirect. Whole larvae transcriptome sequencing and absence of a protein expression data set could confine our predicted functional analysis, as heterogeneity in tissues can complicate the analysis and not all miRNA–mRNA interactions confer mRNA degradation. In addition, miRNA and their targets have to co-localise to exert biological effects. For example, IGFALS is an insulin-like growth factor I (IGF-I) binding partner responsible for increasing the half-life of circulating IGF-I and for preventing possible non-specific metabolic effect of IGF( Reference Boisclair, Rhoads and Ueki 51 ). Reduction of IGFALS reduces circulating IGF-I and, in consequence, growth. IGF-I has been implicated in somatic growth of Atlantic cod( Reference Davie, Porter and Bromage 52 ). In this study, the expression of igfals mRNA had an inverse relationship with miR-9 in the reference feed group (Fig. 2), which showed faster growth( Reference Karlsen, van der Meeren and Rønnestad 23 ). miR-9 is mainly expressed in Atlantic cod brain( Reference Bizuayehu, Johansen and Puvanendran 31 ); however, it has been shown to be expressed in human blood serum( Reference Kong, Zhu and Han 53 ). Although IGFALS is produced in the liver, growth hormone (GH) from the pituitary controls its production. miR-9 can be one of the rheostat of igfals in the GH/IGF axis. Therefore, the role of miR-9 in the accelerated growth of larvae in the reference feed group at stage 2 is either indirect or it does not exist in vivo.
miR-9 also targets phkg1 that activates glycogen phosphorylase, which further catalyses the rate-limiting step in glycogen breakdown. Larvae do not store a lot of glycogen( Reference Conceição, Verreth and Verstegen 54 ), but the reduction in miR-9 expression as development progresses towards the metamorphosis may suggest activation of the glycogenolysis pathway.
We found two targets with inverse expression patterns to that of miR-181a (Table 3, Fig. 2, online Supplementary Fig. S1). Among them, dusp5 is a negative regulator of p38 and stress-activated protein kinases/Jun amino-terminal kinases (SAPK/JNK)( Reference Tanoue, Moriguchi and Nishida 55 ), which are involved in signal transduction pathways with diverse biological functions( Reference Roux and Blenis 56 ). In mammals, miR-181a is implicated in basal activation of T-cell receptor signalling molecules by repressing multiple negative regulators including DUSP5 ( Reference Li, Chau and Ebert 57 ). In addition, mice carrying deletion of some of miR-181 clusters had impaired growth( Reference Henao-Mejia, Williams and Goff 17 ). The significantly higher expression of miR-181a associated with significantly lower expression of dusp5 in the reference feed group as compared with rotifer group (online Supplementary Fig. S1) at stage 2 could result in better growth in the former group through better promotion of cellular differentiation and proliferation by suppressing the repressive action of dusp5. Calml4, one of multiple Ca2+-binding proteins involved in signal transduction, was another miR-181a target differentially expressed in this study. Calml4 is evolutionary conserved, expressed in all eukaryotic cells and involved in signalling pathways of diverse biological processes such as growth, proliferation, metabolism and transport( Reference Chin and Means 58 ). Maternal transcripts of calml4 have been identified in unfertilised eggs of Atlantic cod( Reference Lanes, Bizuayehu and de Oliveira Fernandes 59 ). Hamre et al.( Reference Hamre, Srivastava and Rønnestad 60 ) reported higher content of Ca in rotifers as compared with zooplankton. Therefore, significantly higher expression of calml4 in the aquaculture feed group, as compared with the reference feed group (online Supplementary Fig. S1), could reflect the Ca content in a diet. The elevation of miR-181a expression in the reference feed group at stage 2 may indicate active involvement of miRNA-mediated suppression in the regulation of calml4 expression. However, it is unknown whether calml4, through Ca2+-dependent signalling pathway, has a role in the observed differences in larval performance between the two feeding regimens.
Col1a2 transcript, a component of extracellular matrix secreted by osteoblasts, is a target of miR-19a (Fig. 2). A significant enrichment in col1a2 has been reported in zebrafish( Reference Kessels, Huitema and Boeren 61 ) and sea bass (Dicentrarchus labrax)( Reference Darias, Zambonino-Infante and Hugot 62 ) during larval development. In this study, both feeding groups showed reciprocal expressions of miR-19a and col1a2 at stage 4. In Atlantic cod, ossification occurs in larvae of size 11–20 mm( Reference Pedersen and Falk-Petersen 63 , Reference Herbing, Miyake and Hall 64 ), which corresponds to stage 4 in this study. miR-19a is down-regulated in osteoblast-like cells under peptide-15 stimulation to promote bone formation( Reference Palmieri, Pezzetti and Brunelli 65 ). Although the expression of miR-19a is higher in the reference feed group as compared with the aquaculture feed group at stage 4, its role in Atlantic cod bone formation and its mechanistic action is unknown yet.
miR-206 promotes cell differentiation by regulating transcriptional factor genes that are important in skeletal muscle growth. We predicted that miR-206 targeted a transcriptional factor myb (c-myb). C-myb has been implicated in the proliferation of myoblasts by maintaining their undifferentiated states through inhibition of myogenic differentiation 1 (myoD)( Reference Kaspar, Pajer and Sedlak 66 ). Myogenic regulatory factors (MGF) regulate the transcription of muscle-specific miRNA – for example, myogenic factor 5 (Myf5) and MyoD regulate miR-206 transcription( Reference Sweetman, Goljanek and Rathjen 67 ). At the same time, miR-206 forms a feedback regulatory loop with MGF( Reference Chen, Tao and Li 68 ). Inhibition of miR-206 using antagomiR resulted in a substantially faster growth in Nile tilapia (Oreochromis niloticus)( Reference Yan, Zhu and Guo 69 ). Furthermore, ectopic expression of myoD increased expression of muscle-specific miRNA and induced muscle cell differentiation( Reference Koutsoulidou, Mastroyiannopoulos and Furling 70 ). In Atlantic cod larvae, the expression of myoD1 increased and pax7 decreased after 20 dph( Reference Hamre, Penglase and Rasinger 71 ). The third phase of myogenesis (mosaic hyperplasia), which normally starts by the end of the larval period( Reference Johnston 72 ), contains myogenic precursor cells that give rise to new myotubes( Reference Johnston 72 ). In this study, the inverse relationship between miR-206 and c-myb during metamorphosis in the reference feed group could reflect regulation of the c-myb by miRNA to accommodate the ongoing changes in skeletal muscle. This may suggest that the balance between proliferation and differentiation is important for fast larval growth. Therefore, suppression of c-myb by miR-206 could promote the transcription of myoD, and thereby differentiation of myogenic precursor cells to myotubes, which may imply faster growth.
In this study, we identified differentially expressed miRNA that are known to be involved in different biological pathways. However, our enrichment analysis yielded only one significantly enriched pathway (one carbon pool by folate pathway), which had three miR-206 targets. Moreover, these targets were not differentially expressed between the two groups.
Conclusion
We demonstrate that the effect of the type of start-feed on larval development and growth likely implicates miRNA-mediated regulation. Different types of live feed resulted in differential expression of some miRNA and their putative targets throughout the feeding period. The effects were sometimes extended beyond the first feeding phase. As aquaculture live feeds (rotifers and Artemia) are nutritionally inferior to the reference feed (copepods), a specific enrichment of live feeds and/or feed modifications or supplementation to alter miRNA levels in larvae may offer a strategy to improve larviculture.
Acknowledgements
The study was financed by The Research Council of Norway, projects CODE (no. 199482) and FishmiR (no. 213825). The Research Council of Norway had no role in the design, analysis or writing of this article.
T. T. B. performed SOLiD Sequencing, real-time RT-PCR, analyses and wrote the first draft of the manuscript; T. F. and R. B. E. performed RNA-Seq and contributed to 3′ UTR identification; Ø. K. and T. v. d. M. performed the experiment and sampling; I. R. and K. H. conceived and coordinated the experiment; S. D. J. and I. B. designed and coordinated the study. All the authors contributed to the writing of the manuscript.
The authors declare that there are no conflicts of interest.
Supplementary Material
For supplementary material/s referred to in this article, please visit http://dx.doi.org/doi:10.1017/S0007114516000155








