Introduction
The Phylum Apicomplexa contains a large number of species of protozoan parasites, some of which are recognized as important pathogens that cause human diseases (e.g. Plasmodium spp. and Toxoplasma gondii) or as animal parasites of considerable economic importance (e.g. Eimeria spp. and Neospora spp.) (Kemp et al., Reference Kemp, Yamamoto and Soldati-Favre2013). Apicomplexan parasites have an obligate intracellular existence, and many protozoan parasites also infect the blood cells and organs cells of their vertebrate hosts (Kemp et al., Reference Kemp, Yamamoto and Soldati-Favre2013; Al-Quraishy et al., Reference Al-Quraishy, Abdel-Ghaffar, Dkhil and Abdel-Gaber2021). The group of haemogregarines (Apicomplexa: Adeleorina) is considered a common parasite in reptiles, where Hepatozoon is the most commonly reported parasite. Of note, the gamonts of Hepatozoon display an important life-cycle characteristic as they infect peripheral blood erythrocytes of lizards (Telford, Reference Telford2009).
Hepatozoon species are heteroxenous parasites, implying a complex variety of life cycles involving vertebrate and invertebrate hosts (ticks, mosquitoes, flies). Merogony occurs in the vertebrate host, producing gamonts (without sexual dimorphism) that are found within blood cells. Vertebrate infection is believed to occur through ingestion of infected invertebrate hosts and, in reptiles, transmission usually occurs through the ingestion of an infected vector or following predation of a vertebrate (paratenic host) (Smith, Reference Smith1996; Paperna and Lainson, Reference Paperna and Lainson2004; Al-Quraishy et al., Reference Al-Quraishy, Abdel-Ghaffar, Dkhil and Abdel-Gaber2021).
Given that parasites may infect a wide range of host species, many haemogregarines have been described in Brazilian reptiles, including caiman (Hepatozoon caimani Carini, Reference Carini1909) and snakes (Hepatozoon cevapii O'Dwyer et al., Reference O'Dwyer, Moço, dos S Paduan, Spenassatto, da Silva and Ribolla2013, Hepatozoon massardii O'Dwyer et al., Reference O'Dwyer, Moço, dos S Paduan, Spenassatto, da Silva and Ribolla2013, Hepatozoon cuestensis O'Dwyer et al., Reference O'Dwyer, Moço, dos S Paduan, Spenassatto, da Silva and Ribolla2013, Hepatozoon musa Borges-Nojosa et al., Reference Borges-Nojosa, Borges-Leite, Maia, Zanchi-Silva, da Rocha Braga and Harris2017) (Carini, Reference Carini1909; O'Dwyer et al., Reference O'Dwyer, Moço, dos S Paduan, Spenassatto, da Silva and Ribolla2013; Borges-Nojosa et al., Reference Borges-Nojosa, Borges-Leite, Maia, Zanchi-Silva, da Rocha Braga and Harris2017). In reptiles of the Amazon, Hepatozoon infection has been reported in Boa constrictor (Hepatozoon cf. terzii Paperna and Lainson, Reference Paperna and Lainson2004), Caiman caimani (Hepatozoon caimani Carini, Reference Carini1909) and Ameiva ameiva (Hepatozoon ameivae Carini and Rudolph, Reference Carini and Rudolph1912) (Lainson et al., Reference Lainson, Paperna and Naiff2003b; Paperna and Lainson, Reference Paperna and Lainson2004; Picelli et al., Reference Picelli, da Silva, Ramires, da Silva, Pessoa, Viana and Kaefer2020).
Although the evaluation of morphological values of the host and parasite life-cycle developmental stages is crucial for the correct identification of parasites, molecular data are also essential for differentiating species and genera (O'Donoghue, Reference O'Donoghue2017). Therefore, using morphological and molecular analyses, the present study reports on the presence of a new species of Hepatozoon, Hepatozoon lainsoni sp. nov., with the unique characteristic of parasitizing monocyte/macrophage cells in the A. ameiva lizard.
Materials and methods
Lizards
A total of 111 adult A. ameiva (Lepidosauria, Teiidae) were hand captured at the municipality of Capanema, Pará state, northern Brazil (01°11′37.6″S, 047°10′01.4″). Lizards were maintained as described by Silva et al. (Reference Silva, Diniz, Alberio, Lainson, De Souza and DaMatta2004).
Morphological and morphometric analyses
The lizards were restrained manually, and their blood was collected by cardiac puncture into heparinized (100 U mL−1) 1 mL syringes. The blood smears and the impression smears (imprints) of the lungs, brain, heart, kidney, liver, bone marrow and spleen were air-dried, fixed with absolute methanol and stained with 10% Giemsa Methylene Blue Eosin Merck® diluted in distilled water (pH 7.0) for 50 min, according to Eisen and Schall (Reference Eisen and Schall2000), to allow analysis of the different parasite forms and stages. The samples were examined, and the infected cells were photographed with a Zeiss Axiophot microscope using a 100× immersion objective. Prevalence was estimated as the proportion of infected hosts, expressed as a percentage. The intensity of parasitaemia in blood monocytes was graded according to Silva et al. (Reference Silva, Diniz, Alberio, Lainson, De Souza and DaMatta2004) as negative, low-level infected, medium-level infected and highly infected.
Measurements of the length, width and area of the gamonts and host cells (infected and uninfected) were performed. Morphometric data are presented in micrometres (μm), and for each metric, the averages, ranges and standard deviations were also calculated. Measurements of cells were carried out using a 100× oil immersion objective on a Zeiss Axiophot microscope, calibrated with a stage micrometre. Unstained samples were analysed by differential interference contrast (DIC) microscopy.
Molecular analysis
DNA was extracted from blood using an Illustra blood genomicPrep Mini Spin Kit (GE Healthcare, São Paulo, Brazil) following the manufacturer's instructions. The 18S rDNA gene was amplified with 2 pairs of primers, HepF300/Hep900 (600 bp) primers (Ujvari et al., Reference Ujvari, Madsen and Olsson2004) and 4558/2733 (1000 bp) primers (Mathew et al., Reference Mathew, Van Den Bussche, Ewing, Malayer, Latha and Panciera2000) to detect the Hepatozoon species; reactions were carried out under conditions established by O'Dwyer et al. (Reference O'Dwyer, Moço, dos S Paduan, Spenassatto, da Silva and Ribolla2013) and Netherlands et al. (Reference Netherlands, Cook and Smit2014). To check for possible contamination, nuclease-free water was used as a negative control. A blood sample from a Crotalus durissus terrificus snake that previously tested positive for Hepatozoon spp. was used as the positive control (supplied by the Laboratory of Parasitology of the Institute of Biosciences, UNESP-IB). All reactions were carried out in a Mastercycler pro (Eppendorf, São Paulo, Brazil) thermocycler with the following programme: 94°C for 3 min, 35 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 60 s, followed by a final 7 min extension at 72°C.
The amplification products were subjected to electrophoresis at 80 V in a 1.5% agarose gel, stained with Gel Red, and observed using an ultraviolet transilluminator. The products of interest were purified by adding 2 μL of ExoSAP IT enzyme (GE Healthcare) to 5 μL of each polymerase chain reaction product, according to the manufacturer's recommendations. Amplicons were then sequenced using a 3500 Genetic Analyzer capillary sequencer (Applied Biosystems; Foster City, California, U.S.A) and a BigDye Terminator Cycle Sequencing Ready Reaction Kit v.3.1 (Applied Biosystems; Foster City, California, U.S.A), according to the manufacturer's recommendations. A consensus sequence alignment was performed from the forward and reverse electropherograms using BioEdit software version 7.0.9 (Hall, Reference Hall1999). The sequences obtained (589 and 1052 nt) from A. ameiva (GenBank numbers PP003255 and PP003256) were compared among them and with those of other Hepatozoon isolates available in the GenBank.
Sequences were aligned with the MUSCLE algorithm using Geneious v.7.1.3 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012) for Bayesian inference (BI) and maximum-likelihood (ML) analyses. From this alignment, to compare the 18S rRNA gene in Hepatozoon isolates, a pairwise distance (p-distance) matrix was used (Table 1). For ML phylogeny, JModelTest v.2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012) was used to identify the best evolutionary model. Based on the Akaike information criterion, the TVM + G model was chosen for ML analyses. The phylogeny of the parasite was inferred using PhyML v.3.0 (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010) with 1000 replicate bootstraps (>50%). BI was carried out using MrBayes implemented from the computational resource CIPRES (Miller et al., Reference Miller, Pfeiffer and Schwartz2010), and analysis was run with the nucleotide substitution model GTR + I + G. To search with the Markov chain Monte Carlo method, chains were run with 10 000 000 generations, saving 1 tree every 1000 generations. On the burn-in, the first 25% of generations were discarded, and the consensus trees were estimated using the remaining trees. Bayesian posterior probability cutoff was considered to be >50%. The trees (BI and ML) were visualized and edited using the FigTree v1.3.1 software program (Rambaut, Reference Rambaut2012). The sequences from the phylogenetic tree, their hosts and their GenBank accession numbers are shown in Table 2. The pairwise distance (p-distance) and gene similarity were executed by the MEGA 7 program (Kumar et al., Reference Kumar, Stecher and Tamura2016). A matrix was used to compare the interspecific divergence between the species of Hepatozoon sequences isolated from lizards.
Table 1. Shaded matrix (upper) shows the similarity percentage (%) of the nucleotide sequences and the unshaded matrix (lower) shows the pairwise distance (p-distance) among Hepatozoon spp. isolates from lizards

Sequence length: 1052 nt.
Table 2. Hosts, localities and GenBank accession numbers for the 18S rDNA sequences of Hepatozoon spp., Haemogregarina spp. and Hemolivia spp. used in the phylogenetic analyses (except for the sequence from this study and outgroup)

Results
Of the 111 A. ameiva lizards screened, 55 (49.5%) showed positivity for haemoparasites infecting monocytes in the peripheral blood (Figs 1 and 2) and organ imprints (Fig. 3). The level of infection was usually low (up to 2 parasites per microscopic field), although 5 (9.1%) of the lizard population presented high-level parasitaemia (above 3 parasites per microscopic field). Developmental stages were observed in the spleen (Fig. 3) and bone marrow (not shown), but not detected in other organs. Through morphological, morphometric and molecular evaluation, a previously undescribed species of haemogregarine parasite was identified; this species belongs to the genus Hepatozoon Miller, 1908 (Adeleorina: Hepatozoidae).
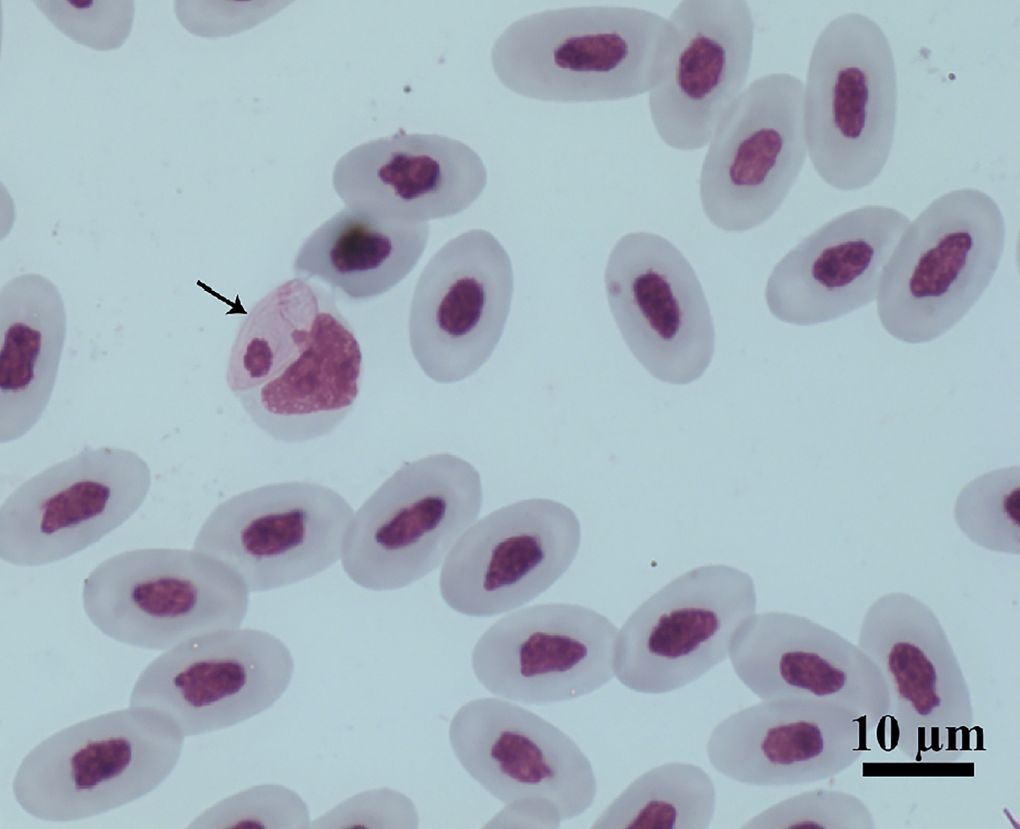
Figure 1. Intramonocytic gamonts (arrows) of Hepatozoon lainsoni sp. nov. from peripheral blood of the lizard Ameiva ameiva stained with Giemsa. Scale bar: 10 μm.
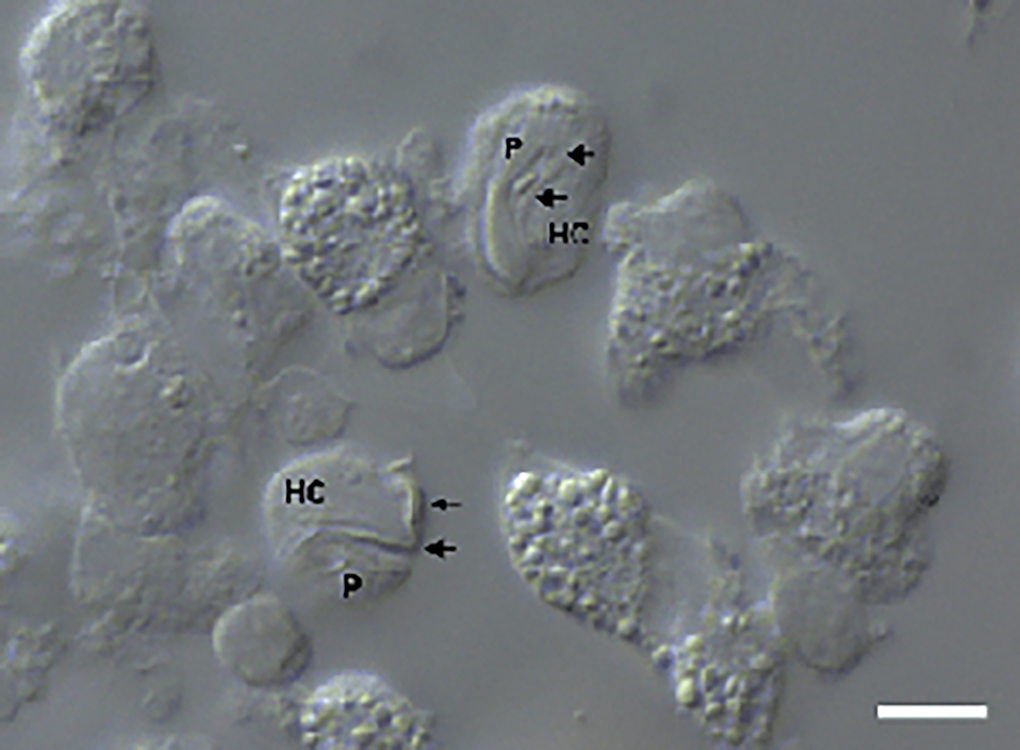
Figure 2. Intramonocytic gamonts (arrows) of H. lainsoni sp. nov. from peripheral blood of the lizard A. ameiva observed by DIC microscopy. HC, host cell; P, parasite. Scale bar: 10 μm.
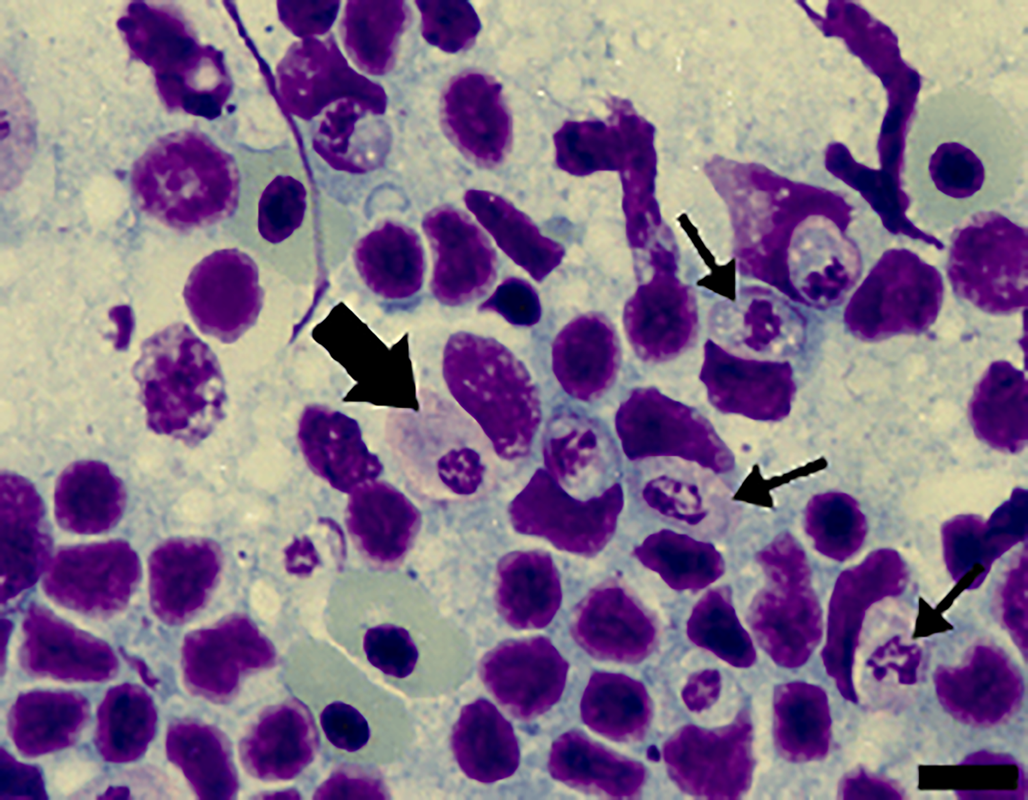
Figure 3. Cells from the spleen imprints stained with Giemsa from A. ameiva lizards infected with the parasite. Immature gamonts (arrows) and mature gamonts (big arrow). Scale bar: 10 μm.
Species description
Phylum Apicomplexa Levine, 1970
Class Conoidasida Levine, 1988
Subclass Coccidia Leuckart, 1879
Order Eucoccidiorida Léger, 1911
Suborder Adeleorina Léger, 1911
Family Hepatozoidae Wenyon, 1926
Genus Hepatozoon Miller, 1908
Hepatozoon lainsoni sp. nov. Morais and Silva, 2023
Taxonomic summary
Type-host: Ameiva ameiva (Linnaeus, 1758) (Squamata: Teiidae: Teiinae).
Type locality: In areas of the Capanema municipality, Pará state, northern Brazil (01°11′37.6″S, 047°10′01.4″W).
Site of infection: Monocytes and macrophages from peripheral blood, spleen and bone marrow, respectively.
Vector: Remains unknown; we speculate that it is an external blood-sucking vector, such as a tick species.
Etymology: The species is named after the parasitologist, Ralph Lainson, who extensively researched the protozoan parasites of the Brazilian Amazon, and who described this parasite in a preliminary note (Lainson et al., Reference Lainson, De Souza and Franco2003a).
Prevalence: Of the 111 lizards analysed, 55 (49.5%) were positive.
Material deposited: Hapantotypes, 1 blood smear, 1 imprint of the spleen and 1 imprint of bone marrow from the A. ameiva lizard were deposited in the collection of the National Institute of Amazonian Research (INPA), Manaus, Brazil [INPA 24].
Gene sequence: The 18S rRNA gene sequences (589 and 1052 bp), obtained from the blood of A. ameiva, were deposited in the GenBank, under accession numbers PP003255 and PP003256.
Morphological and morphometrical analysis
Gamonts: Gamonts were observed exclusively in monocytes of infected lizards. No division stages were detected in blood films. Gamonts were oval and conspicuous in the monocyte cytoplasm, averaging 10.0–13.5 × 4.5–7.7 (12.0 ± 0.8 × 5.9 ± 0.6) μm2 (n = 50) in size (Table 3), and with an area of 36.9–74.1 (57.9 ± 8.2) μm2 (n = 50). The parasite cytoplasm was stained greyish-blue or pale pink with thin visible reddish staining at the membrane. Some gamonts contained numerous magenta-staining granules. Nuclei were small, averaging 1.9–5.3 × 1.5–4.0 (3.6 ± 0.8 × 2.6 ± 0.5) μm2 (n = 50) in size, with an area of 3.2–11. (6.9 ± 1.7) μm2 (n = 50); they were rounded or irregular shaped, demonstrated dark purple staining and were located at 1 end of the parasite (Fig. 1). DIC microscopy revealed a thin body, with a prolongation, that was doubled on itself and with a nucleus situated in the middle of the parasite (Fig. 2).
Table 3. Measurements (μm) of gamonts of Hepatozoon lainsoni sp. nov. and Hepatozoon sp., infecting lizards from Brazil
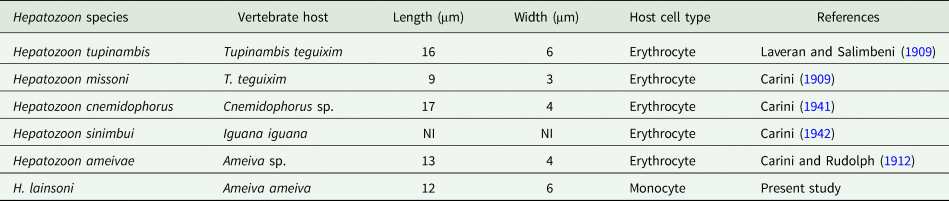
NI, not informed.
Host cells: Host cells were enlarged. Parasitized monocytes measured 11.3–20.9 × 8.9–16.0 (14.9 ± 2.1 × 12.5 ± 7.0) μm2 (n = 50) in size, with an area of 87.2–225.6 (146.2 ± 30.9) μm2 (n = 50). Nuclei of parasitized monocytes measured 7.3–14.3 × 2.4–8.9 (10.7 ± 1.6 × 5.6 ± 1.4) μm2 (n = 50) in size, with an area of 32.5–88.4 (48.7 ± 10.2) μm2 (n = 50). Gamonts displace the nuclei laterally, occasionally becoming deformed and flattened by the parasite. Numerous granules in the cytoplasm were occasionally seen. Uninfected monocytes measured 9.6–17.2 × 8.3–15.3 (13.8 ± 1.6 × 12.2 ± 1.5) μm2 (n = 50) in size, with an area of 67.6–186.3(139.3 ± 24.8) μm2 (n = 50); and nuclei measured 7.8–12.3 × 4.4–10.3 (10.2 ± 1.1 × 7.4 ± 1.4) μm2 (n = 50) in size, with an area of 31.6–86.4 (61.8 ± 13.2) μm2 (n = 50).
Tissue developmental stages (mature and immature gamonts): Tissue stages in the spleen and bone marrow were seen. Sometimes macrophages of the spleen presented 2 parasites in a single cell, with very abundant forms seen in the spleens of some infected lizards (Fig. 3). Spleen and bone marrow of some infected lizards showed immature gamonts, characterized by an elliptical shape, which measured 7.4–9.7 × 5.6–6.9 (8.3 ± 0.8 × 6.4 ± 0.4) μm2 (n = 10), and with an area of 29.0–51.0 (43.6 ± 7.4) μm2 (n = 10). These gamonts had purplish or pinkish cytoplasm with bright granules and fragmented nuclei that occupied the central position or were displaced towards one of the extremities. Another gamont form presented an oval body measuring 8.4–13.5 × 4.4–6.4 (11.3 ± 1.7 × 5.5 ± 0.7) μm2 (n = 10) and with an area that measured 35.2–63.6 (47.7 ± 8.4) μm2 (n = 10); this form had pinkish cytoplasm, with rounded or fragmented nuclei that were displaced towards one of the extremities of the parasite, representing mature gamonts. These stages have variable sizes (Fig. 3).
Morphological and molecular analyses identify a new species of Hepatozoon (Apicomplexa: Adeleorina: Hepatozoidae).
Remarks
Prior to this study, Hepatozoon spp. were reported infecting erythrocytes of reptiles, and occasionally leucocytes. The haemogregarine described herein appears to infect monocytes of the peripheral blood and division stages were seen only in internal organs. Gamonts of H. lainsoni sp. nov. presented differences in morphological and morphometrical characteristics at the blood and tissues stages, when compared to other species of Hepatozoon described in A. ameiva of Minas Gerais and the Amazonas states in Brazil (Carini and Rudolph, Reference Carini and Rudolph1912; Picelli et al., Reference Picelli, da Silva, Ramires, da Silva, Pessoa, Viana and Kaefer2020), or any other species currently infecting lizard hosts (Table 4).
Table 4. Measurements (μm) of host cells and gamonts of H. lainsoni sp. nov. observed in infected A. ameiva lizards from Pará state, Brazil
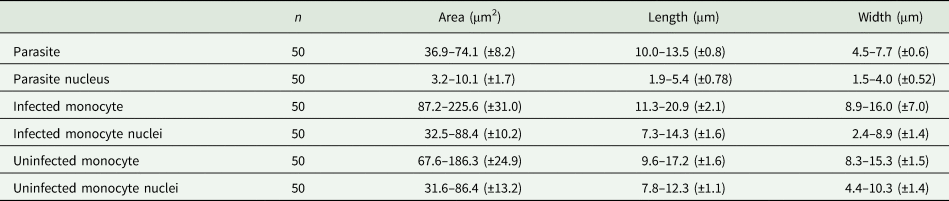
±, standard deviation.
Erythrocytic haematozoans of A. ameiva were described by Lainson et al. (Reference Lainson, De Souza and Franco2003a) to be slim parasites measuring 13.1 × 3.0 μm2 and with a nucleus located at the parasite's curved extremity, leading the authors to compare these with haematozoans of H. ameivae from Minas Gerais, Brazil (Carini and Rudolph, Reference Carini and Rudolph1912). In 2020, Picelli et al. redescribed H. ameivae in the peripheral blood of the same host in the Central Amazon by morphological, morphometrical and phylogenetic analyses. Gamonts were found infecting erythrocytes. The parasites were described to be measuring 14.2 × 4.5 μm2, with curved extremities and with nuclei that were rounded or irregular in shape, and measuring 4.9 × 3.2 μm2.
With regards to other species from host lizards, H. lainsoni sp. nov. have a similar size range to those of H. caimani Carini, Reference Carini1909 (12.15 × 4.3 μm2) from the caiman Caiman c. crocodilus (Lainson et al., Reference Lainson, Paperna and Naiff2003b), Hepatozoon terzii Sambon and Seligman, 1907 (12.3 × 4.3 μm2) from snake B. constrictor (Paperna and Lainson, Reference Paperna and Lainson2004), Hepatozoon quagliattus Úngari, Netherlands, Silva and O'Dwyer, 2021 (13.58 × 6.22 μm2) from the sleep snake Dipsas mikanii (Úngari et al., Reference Úngari, Netherlands, de Alcantara, Emmerich, da Silva and O'Dwyer2021) and are smaller than H. musa Borges-Nojosa, Borges-Leite, Maia, Zanchi-Silva, da Rocha Braga and Harris, Reference Borges-Nojosa, Borges-Leite, Maia, Zanchi-Silva, da Rocha Braga and Harris2017 (18.9 × 3.8 μm2) from snake Philodryas nattereri (Borges-Nojosa et al., Reference Borges-Nojosa, Borges-Leite, Maia, Zanchi-Silva, da Rocha Braga and Harris2017).
The new species, H. lainsoni sp. nov., is characterized by elongated gamonts, which appear to have an oval shape and are not bent at the end. Gamonts were surrounded by a delicate capsule that increased the size of the host cell. Additionally, H. lainsoni sp. nov. have small nuclei that are located at 1 end of the parasite. Hepatozoon lainsoni sp. nov. share certain characteristics with H. caimani, since the gamonts doubled by themselves, and had a visible capsule where both ends were rounded. However, H. caimani exhibit gamont nuclei that are located laterally in the parasite, and extracellular parasites can be seen occasionally in the slim form.
In contrast to H. lainsoni sp. nov., gamonts of H. musa are longer and thinner, and are curved at both ends with their nuclei in the central position. Mature gamonts of H. quagliattus are elongated with a thin parasitophorous vacuole and are slightly curved at 1 end; the gamont nucleus is larger than that of H. lainsoni sp. nov. and slightly displaced to 1 side of the parasite.
With regards to tissue stages, H. lainsoni sp. nov. exhibit immature and mature gamonts in the spleen and bone marrow, with high parasitaemia in the spleen macrophages of some lizards. These gamonts presented a rounded body, colourless cytoplasm and were variable in size.
Molecular analysis
Both the newly amplified sequences from this study have shown 100% similarity among them. A large sequence (1052 nt) was used to construct BI and ML trees. The trees resulted in identical topologies with supported node value clades (Fig. 4). The trees are formed by haemogregarine isolates, haemogregarine (Haemogregarinidae, Karyolysidae and Hepatozoidae) and haemococcidia isolates as an outgroup. The main clade was subdivided into 6 subclades. The 1st subclade (clade I) comprises Hepatozoon isolates from reptiles, anurans and marsupials, including the isolate from this study; furthermore, the isolate from this study grouped together with 3 isolates, 2 from marsupial hosts (FJ719813/FJ719814) and 1 from lizards (KM234614). The 2nd subclade (clade II) comprises Hepatozoon isolates from mammals; the 3rd subclade (clade III) describes isolates from Karyolysus and Hepatozoon species from lizards; the 4th subclade (clade IV) presents Haemolivia isolates; the 5th subclade (clade V) comprises Dactylosoma isolates from anurans and the 6th (clade VI) comprises Haemogregarina isolates from turtles.

Figure 4. Phylogenetic tree of Hepatozoon spp. based on 18S rRNA region gene (1052 bp – sequence length). The topology was identical across ML and BI with supported node value clades.
Discussion
Species of Hepatozoon have a wide geographic distribution, infecting amphibians, reptiles, birds and mammals. To date, species of Hepatozoon have been reported mainly infecting erythrocytes of reptiles. However, other species infecting mammals and birds are found mainly infecting leucocytes (Smith, Reference Smith1996; Godfrey et al., Reference Godfrey, Nelson and Bull2011; Ebani and Mancianti, Reference Ebani and Mancianti2022). This study provides molecular analysis and morphological and morphometric descriptions of the blood forms and tissue stages of H. lainsoni sp. nov., a species that infects monocytes of the lizard A. ameiva.
In the present investigation, blood smears from infected lizards usually showed low parasitaemia. A previous study analysing blood from A. ameiva reported only 10% of the lizard population with high parasitaemia and a higher relative increase in the population of blood monocytes, many of which were infected with H. lainsoni sp. nov. (Lainson et al., Reference Lainson, De Souza and Franco2000a; Bonadiman et al., Reference Bonadiman, Miranda, Ribeiro, Rabelo, Lainson, Silva and DaMatta2010). Similarly, Úngari et al. (Reference Úngari, Santos, O'Dwyer, da Silva, Rodrigues Santos, da Cunha, de Melo Costa Pinto and Cury2018) observed low-level parasitaemia in captive snakes from Brazil with positive blood smears. Other studies of Hepatozoon sp. in reptiles reported 0.25% parasitaemia in the sleep snake D. mikanii (from Goiás state, Brazil) when infected with H. quagliattus Úngari and O'Dwyer, 2021 (Úngari et al., Reference Úngari, Netherlands, de Alcantara, Emmerich, da Silva and O'Dwyer2021) and 0.73% parasitaemia in A. ameiva from Central Amazonia infected with H. ameivae Carini and Rudolph, Reference Carini and Rudolph1912 (Picelli et al., Reference Picelli, da Silva, Ramires, da Silva, Pessoa, Viana and Kaefer2020).
The overall prevalence in this study was 49.5%, which is close to that reported in the preliminary note about this parasite infecting A. ameiva (41.7%) (Lainson et al., Reference Lainson, De Souza and Franco2003a). The prevalence of Hepatozoon sp. infection observed in our study is in agreement with another study of A. ameiva infected with H. ameivae Carini and Rudolph, Reference Carini and Rudolph1912 from Brazil, which found 55.5% of prevalence in 72 lizards in the Amazonas state (Picelli et al., Reference Picelli, da Silva, Ramires, da Silva, Pessoa, Viana and Kaefer2020). Furthermore, a higher prevalence of the occurrence of Hepatozoon sp. infecting free-ranging caimans (C. crocodilus yacare) was reported in a study in the state of Mato Grosso, Brazil (70.8%) (Bouer et al., Reference Bouer, André, Gonçalves, Luzzi, De Oliveira, Rodrigues, Varani, De Miranda, Perles, Werther and Machado2017) and in snakes (Python regius) in Ghana (78.2%) (Sloboda et al., Reference Sloboda, Kamler, Bulantová, Votýpka and Modrý2007), both using blood smear examination. Lower prevalences of 2–17.6% were observed in lacertid hosts from the Maghreb region of North Africa (Maia et al., Reference Maia, Harris and Perera2011) and in Asian snakes (22.2%) (Haklová et al., Reference Haklová, Majláthová, Majláth, Harris, Petrilla, Litschka-Koen, Oros and Peťko2014). Although different methodologies were used in these studies, Maia et al. (Reference Maia, Harris and Perera2011) suggested that infection with Hepatozoon is age-related, considering that the prevalence among snakes seems to be higher than among lizards, and that snakes live much longer than lizards, so that age is related to a higher chance of acquiring the infection, which might persist for a long time (Jakes et al., Reference Jakes, O'Donoghue and Whittier2003; Javanbakht et al., Reference Javanbakht, Široký, Mikulíček and Sharifi2015).
To date, species of Hepatozoon have been reported as infecting the erythrocytes and leucocytes of reptile, mammal and bird hosts (Godfrey et al., Reference Godfrey, Nelson and Bull2011; O'Donoghue, Reference O'Donoghue2017; Ebani and Mancianti, Reference Ebani and Mancianti2022). In this study, we further characterized a species of Hepatozoon infecting monocytes and macrophages. Morphological and morphometric characteristics and phylogenetic analysis distinguished this species from other Hepatozoon species of lizards, such as H. ameivae Carini and Rudolph, Reference Carini and Rudolph1912, Hepatozoon tupinambis Laveran and Salibeni, 1909, Hepatozoon missoni Carini, Reference Carini1909, Hepatozoon cnemidophorus Carini, Reference Carini1941 and Hepatozoon sinimbui Carini, Reference Carini1942 (Carini, Reference Carini1909, Reference Carini1941, Reference Carini1942; Laveran, 1909). Through phylogenetic analysis, it was possible to observe that the Hepatozoon species from this study was grouped closer to Hepatozoon from marsupial hosts than lizard hosts. This fact corroborates with the observed morphological data. Hepatozoon lainsoni was found infecting defence cells, a characteristic normally found in mammals; while in reptiles it is common to be found in erythrocytes. Therefore, the morphological and molecular data suggest that H. lainsoni may have an ancestor closer to mammals than to reptiles.
The morphometric and morphological analysis showed that the average size and tissue stages of H. lainsoni sp. nov. closely resembled those previously described by Lainson et al. (Reference Lainson, De Souza and Franco2003a). Gamonts of Hepatozoon from this study revealed differences compared to the other species of Hepatozoon described in reptiles. In the literature, the Hepatozoon species described in A. ameiva, H. ameivae Carini and Rudolph, Reference Carini and Rudolph1912 are recognized to have a slim and elongated form, with both rounded and slightly arched extremities, with nuclei located at its curved extremity, and to infect erythrocytes. In contrast, H. lainsoni gamonts had an oval shape with rounded extremities and a small nucleus and infected monocytes. Indeed, DIC microscopy revealed the gamont to have a slim body. These data were supported by ultrastructural analysis of the blood forms, which showed an elongated form, with some embracing the monocyte cytoplasm and, sometimes in intimate contact with the host cell nucleus, suggesting a type of nutrient capture (Silva et al., Reference Silva, Diniz, Alberio, Lainson, De Souza and DaMatta2004). Again, these results corroborate the description of a new species of Hepatozoon.
One noticeable characteristic of the Hepatozoon species was the alterations in the monocyte host cell. In the present study, enlargement and deformation of the nucleus, which was flattened by the parasite, were observed. Similar morphological alterations in the host monocyte after parasite infection were previously observed by Lainson et al. (Reference Lainson, De Souza and Franco2003a) and Silva et al. (Reference Silva, Diniz, Alberio, Lainson, De Souza and DaMatta2004) using optical and transmission electron microscopy, respectively. Furthermore, accumulation and association of mitochondria and Golgi complex vesicles at the intracellular parasite were also observed by Silva et al. (Reference Silva, Diniz, Alberio, Lainson, De Souza and DaMatta2004). Shape alteration, lateral displacement of the nucleus and enlargement of infected blood cells have already been reported in snakes Hydrodynastes gigas and C. terrificus infected by Hepatozoon sp. in Brazil (Moço et al., Reference Moço, O'Dwyer, Vilela, Barrella and Da Silva2002). In the present study, tissue stages were observed in the spleen and bone marrow, with the presence of immature and mature gamonts in some lizards; merogony and cystic forms were not seen. Indeed, we did not examine the skeletal muscle and intestines of infected lizards, where the occurrence of merogony and tissue cysts containing cystozoites may be possible, as seen in the host cells of mammal hosts infected with Hepatozoon felis, Hepatozoon americanum and Hepatozoon silvestris (Hodžić et al., Reference Hodžić, Alić, Prašović, Otranto, Baneth and Duscher2017).
Dividing meronts were observed in the lungs and/or liver tissues of reptiles infected with H. terzii (Paperna and Lainson, Reference Paperna and Lainson2004), Hepatozoon kisrae (Paperna et al., Reference Paperna, Kremer-Mecabell and Finkelman2002) and H. quagliattus (Úngari et al., Reference Úngari, Netherlands, de Alcantara, Emmerich, da Silva and O'Dwyer2021). Conversely, Lainson et al. (Reference Lainson, Paperna and Naiff2003b) described H. caimani Carini, Reference Carini1909 in Caiman c. crocodilus and detected meronts only in the lamina propria of experimentally infected caimans, suggesting that merogony is limited to the lamina propria of the small intestine and justifying the failure of other authors to detect meronts in the viscera of naturally infected caimans. It is probable that a similar pattern of merogonic stages may occur in H. lainsoni and further investigative endeavours are needed to clarify this.
Therefore, these discoveries underscore the need for expanded research on haemoparasite infections in lizards and other reptiles, especially in the Brazilian Amazon, which represents a geographic region with a great diversity of hosts and the possibility of finding genetically distinct haemogregarines.
Data availability statement
Representative sequences related to this article have been deposited in the GenBank under accession numbers PP003255 and PP003256.
Acknowledgements
We are grateful to Instituto Evandro Chagas/SVSA/MS, FADESP and PROPESP-UFPA.
Author's contribution
All the authors of this manuscript contributed substantially with the following activities: R. A. P. B. M., E. O. S., J. A. P. D., W. S. and A. P. D. R.: Study design, data collect, analysis and interpretation. L. P. U. and L. H. O.: performed molecular and statistical analyses. R. A. P. B. M. and E. O. S.: Manuscript writing and literature review. R. A. D. and E. O. S.: Manuscript review and final approval for article submission version.
Financial support
This work was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (E. O. S., grant number 478119/2006-2); PROPESP/UFPA (Qualified Publication Support Program) (E. O. S., grant number 02/2022); Coordenação de Pessoal de Nível Superior (CAPES)/PROCAD CAPES NF/2009 (R. A. P. B. M., grant number 21/2009) and FAPESP (LPU grant number 2018/00754-9; 2018/09623-4).
Competing interests
None.
Ethical standards
Experiments comply with Brazilian animal protection laws (IBAMA doc. 02018.000301/02-13, MMA SISBIO No. 12420-2). All experimental procedures were approved by the Animal Ethics Committee of the Evandro Chagas Institute (CEUA No. 13/2019-IEC).