INTRODUCTION
Human parechoviruses (HPeVs) are small non-enveloped RNA viruses with a single-stranded genome of positive polarity that is ~7·3 kb in length. HPeVs are members of the genus Parechovirus of the family Picornaviridae [Reference Pallansch, Oberste, Whitton, Knipe and Howley1]. Based on genotypic analyses, 17 HPeV genotypes have been defined: HPeV type 1 (HPeV1) to HPeV17 [Reference Pallansch, Oberste, Whitton, Knipe and Howley1, Reference Chuchaona2]. HPeV infection can be associated with a wide variety of clinical manifestations, ranging from asymptomatic infections or mild disease to severe disease symptoms. HPeV1, 3, and 6 frequently cause mild diseases such as gastroenteritis, respiratory infections, and rash in infants and children [Reference Wildenbeest3, Reference Benschop4]. In addition, HPeV3 causes severe diseases such as sepsis and meningoencephalitis in neonates and young infants [Reference Aizawa5]. HPeV3 infection in young infants has been reported globally, including Europe [Reference Schuffenecker6, Reference Piralla7], North America [Reference Walters8, Reference Felsenstein9], Asia [Reference Ghanem-Zoubi10–Reference Shoji12], and Australia [Reference Cumming13].
HPeV3 is reportedly a causative agent of epidemic myalgia and myositis in children and in adults [Reference Yamamoto14–Reference Mizuta16]. HPeV3 seropositivity may thus help to explain why myalgia and myositis occur in child and adult populations. HPeV3 seropositivity in children was low [Reference Ito17, Reference Westerhuis18]. There are no data on HPeV3 seropositivity in adults aged >40 years [Reference Ito17, Reference Westerhuis18]; thus it is important to clarify HPeV3 seropositivity in adults. We hypothesized that adults may have increased susceptibility to HPeV3 due to a lack neutralizing antibodies. Furthermore, it is also important to detect phases of obtaining antibodies to HPeV1, 3, and 6 in children to provide new seroepidemiological insight into HPeVs infection.
In Japan, there is nationwide pathogen surveillance that is based on the Infectious Diseases Control Law [19]. The category I–V infectious diseases mainly caused by bacteria or viruses are targeted in pathogen surveillance. Laboratory examination is performed at prefectural public health institutes using a conventional method for culture and/or a method for amplifying a gene. We have isolated HPeVs using a cell culture method at Niigata Prefectural Institute of Public Health and Environmental Sciences and 120 HPeV strains as of 2014 have been isolated from clinical specimens from patients with different disease conditions.
In this study, we clarify why myalgia occurs in an adult population by investigating HPeV seropositivity in different age groups. Furthermore, we characterize HPeVs infection from the aspects of seropositivity and epidemiology.
MATERIAL AND METHODS
Serum samples
Between July and September 2010, a total of 259 serum samples were collected from participants in the study. The participants were patients who consulted a paediatrician and Niigata Prefecture office staff who had a health check-up. The overall age distribution of enrolled individuals was between 2 months and 61 years. Informed consent was obtained from all participants or their guardians prior to their participation in the study. The samples were divided on the basis of participants' age groups: 2–6 months, 7–11 months, 1 year, 2 years, 3 years, 4 years, 5 years, 5-year age groups from 6–40 years, and 10-year age groups from 41–61 years. In order to detect a phase of obtaining antibody to HPeV1, 3, and 6 in children in detail, the younger age groups were divided into a narrower range than those of the older age groups.
Neutralization test
All serum samples were stored at –20 °C and then inactivated at 56 °C for 30 min before testing. Three prototype strains (HPeV1: Harris, HPeV3: A308/99, and HPeV6: NII561-2000) were used as an antigen (i.e. challenge virus), and standard micro-neutralization testing was performed. Twenty-five microlitres of 100 median tissue culture infective dose (TCID50) viruses and 25 μl of serially diluted sera (twofold serial dilutions from 1:10 to 1:2048) were mixed in a 96-well plate and incubated at 37 °C for 1 h. Subsequently, 100 μl of suspended LLC-MK2 cells (106/ml) were added to the wells. The plate was then placed in a CO2 incubator at 37 °C for 10 days, and the appearance of a cytopathic effect (CPE) was monitored from day 3 using light microscopy. Challenge virus titre was confirmed as containing 100 TCID50 by back titration. Neutralizing antibody titres (NATs) were expressed as the reciprocal of the highest serum dilution that inhibited CPE by 50%. NATs exceeding 1:10 were considered to be positive.
Virus isolation and identification
Between 1997 and 2014, 19155 clinical specimens (mainly stool, respiratory swab, and cerebrospinal fluid) were collected from patients with viral infection for the nationwide pathogen surveillance in Japan, which is based on the Infectious Diseases Control Law [19]. Most of clinical specimens were collected by physicians and paediatricians. In Niigata Prefectural Institute of Public Health and Environmental Sciences, the virus isolation method has been unchanged since 1987. A combination of cell lines used for virus isolation, however, had minor changes. Six adherent cell lines, MDCK, Caco-2, RD-18S, Vero9013, LLC-MK2, and A549, were used most recently. Cell lines and culture conditions have been previously described [Reference Watanabe20]. When CPE was observed, virus identification was performed using immunofluorescence assay, neutralization test, haemagglutination inhibition test, polymerase chain reaction (PCR), reverse transcription–PCR (RT–PCR), or sequence analysis.
HPeV identification and typing
Viral RNA was extracted from isolates using EXTRAGEN II (Tosoh, Japan), and cDNA was then synthesized using SuperScript VILO Master Mix (Invitrogen, USA). An RT–PCR assay was used to amplify the VP1 region [Reference Ito21]. The primers used for PCR performed in a GeneAmp® PCR system 9700 thermal cycler (Applied Biosystems, USA) were as follows: HPeV-VP1S forward primer (5′-GGD ARR MTK GGD VAW GAY GC-3′) and HPeV-VP1AS reverse primer (5′-CCA TAR TGY TTR TAR AAA CC-3′). The cycling conditions included 30 cycles of 98 °C for 10 s, 42 °C for 30 s, 72 °C for 1 min; the amplicon size was 830 bp. PCR products were purified using AMPure XP (Beckman Coulter, USA). After a dye terminator cycle sequencing reaction using the BigDye Terminator v. 3.1 Cycle Sequencing kit (Applied Biosystems), the dye terminator cycle sequencing reaction mixtures were purified using CleanSEQ (Beckman Coulter) and sequenced using an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). The VP1 sequence data were analysed using sequencer software. Multiple sequence alignment was performed using Clustal W. The distance matrix was calculated using the maximum composite likelihood method, and a phylogenetic tree was constructed by the neighbour-joining method using Molecular Evolutionary Genetic Analysis software (MEGA version 6) [Reference Tamura22]. Phylogenetic trees were constructed using complete VP1 sequences obtained in this study, along with others from the DDBJ/ENA/GenBank database. The DDBJ/ENA/GenBank accession numbers for VP1 gene sequences determined in this study are AB900164–AB900166 and LC062393–LC062507.
Statistical analyses
Statistical analyses were performed using the IBM SPSS Statistics 23·0 package (IBM Corp., USA). Neutralizing antibody seroprevalences in HPeV genotypes were compared using χ 2 test. Kruskal–Wallis test was used to determine the possible statistically significant differences in neutralizing antibody geometric mean titres (GMTs) among HPeV genotypes, and Mann–Whitney U test was further used to compare between HPeV genotypes. In the calculations, NAT < 1:10 was regarded as 1 and NAT >1:2560 as 2561. Two-tailed P < 0·05 was considered to indicate statistical significance.
RESULTS
HPeV seropositivity
Seroprevalences of and GMTs to HPeV1, 3, and 6 increased with age, and HPeV1 showed increased seropositivity at younger ages, followed by HPeV3 and HPeV6. Seroprevalences of and GMT to HPeV1 peaked at 90·9% and 251·7 in the 3 years age group, those of HPeV3 peaked at 80·0% and 345·6 in the 5 years age group, and those of HPeV6 peaked at 90·0% and 304·1 in the 11–15 years age group, respectively (Table 1). After reaching maximum levels, seroprevalence of and GMTs to these genotypes were maintained at approximately 80% and 100, respectively. However, HPeV3 seropositivity began to decrease in the 36–40 years age group. In particular, compared to HPeV1 and HPeV6 seropositivity, HPeV3 seropositivity in subjects aged >40 years was significantly lower (P < 0·001, P = 0·003).
Table 1. Seroprevalences and geometric mean titres against HPeV neutralizing antibodies in Niigata, Japan, between July and September 2010
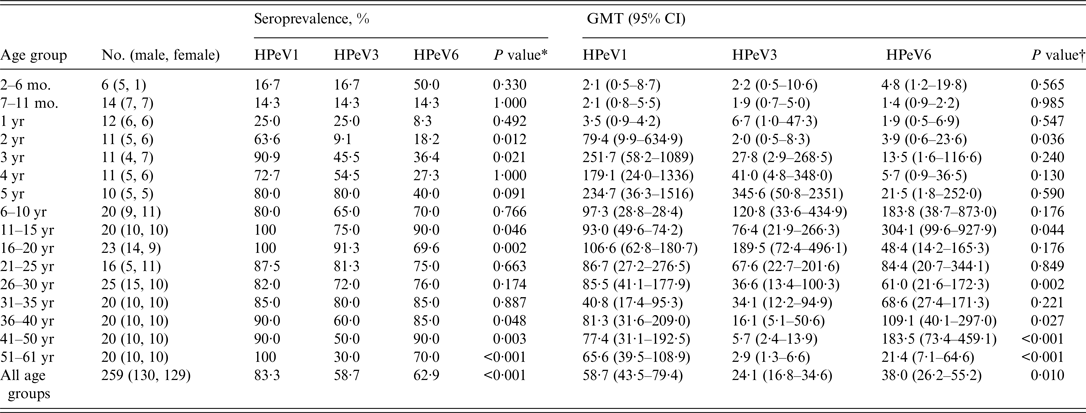
mo., Months; yr, years; HPeVs, human parechoviruses; GMT, geometric mean titre; CI, confidence interval.
* χ 2 test.
† Kruskal–Wallis test.
Significant differences in seroprevalences of and GMTs to HPeV1, 3, and 6 were observed (Table 1). Notably, for seroprevalences of and GMTs to HPeV1 and HPeV3, significant differences were observed in groups aged 41–50 years (P = 0·006) and 51–61 years (P < 0·001) and in those aged 41–50 years (P < 0·001) and 51–61 years (P < 0·001), respectively. As for seroprevalences of and GMTs to HPeV3 and HPeV6, significant differences were observed in groups aged 41–50 years (P = 0·006) and 51–61 years (P = 0·011) and in those aged 41–50 years (P < 0·001) and 51–61 years (P = 0·010), respectively.
HPeV yearly distribution
HPeV strains were isolated from the clinical specimens every year between 1997 and 2014, except for 2002 and 2003 (Fig. 1). According to data in the Infectious Agents Surveillance Report [23], HPeVs were reported in only 22 and 18 cases in Japan in 2002 and 2003, respectively; therefore we were unable to isolate HPeV strains in Niigata. A total of 118 HPeV strains were isolated from 66 stool samples, 44 respiratory swabs, and eight cerebrospinal fluid samples. HPeV strains were isolated from eight cell lines (LLC-MK2, Vero, Vero9013, BSC-1, Caco2, RD18S, HEp2, HeLa), and frequently plural cell lines. LLC-MK2 cells (83/118) were the most sensitive, the second most sensitive cell line was Caco2 cells (37/118), and the third was Vero cells (34/118). The isolated number of strains was as follows: HPeV1 (55, 47%); HPeV3 (50, 42%), and HPeV6 (13, 11%). The sensitivity of the cell lines to the genotypes varied. LLC-MK2 cells (39/55) and Caco2 cells (28/55) to HPeV1, LLC-MK2 cells (42/50) and Vero cells (16/50) to HPeV3, and Caco2 cells (6/13) and Vero cells (4/13) to HPeV6 indicated more sensitivity. HPeV1 was more frequently isolated than the other two genotypes (Fig. 1). HPeV1 infection endemically prevailed in Niigata, and HPeV1 isolate numbers in a year tended to increase from 2012 onwards. HPeV3 was also frequently isolated in Niigata before 2008; however, its circulation changed after 2008, and HPeV3 was isolated every 3 years, in 2008, 2011, and 2014 (Fig. 1). An HPeV3 epidemic occurred in Japan in those 3 years [Reference Aizawa5, 23, 24]. HPeV6 was infrequently isolated and HPeV6 infection was sporadic; local outbreaks of HPeV6 occurred in 2000 and 2001 (Fig. 1) [Reference Watanabe20]. HPeV1, 3, and 6 infections thus showed different epidemic patterns.
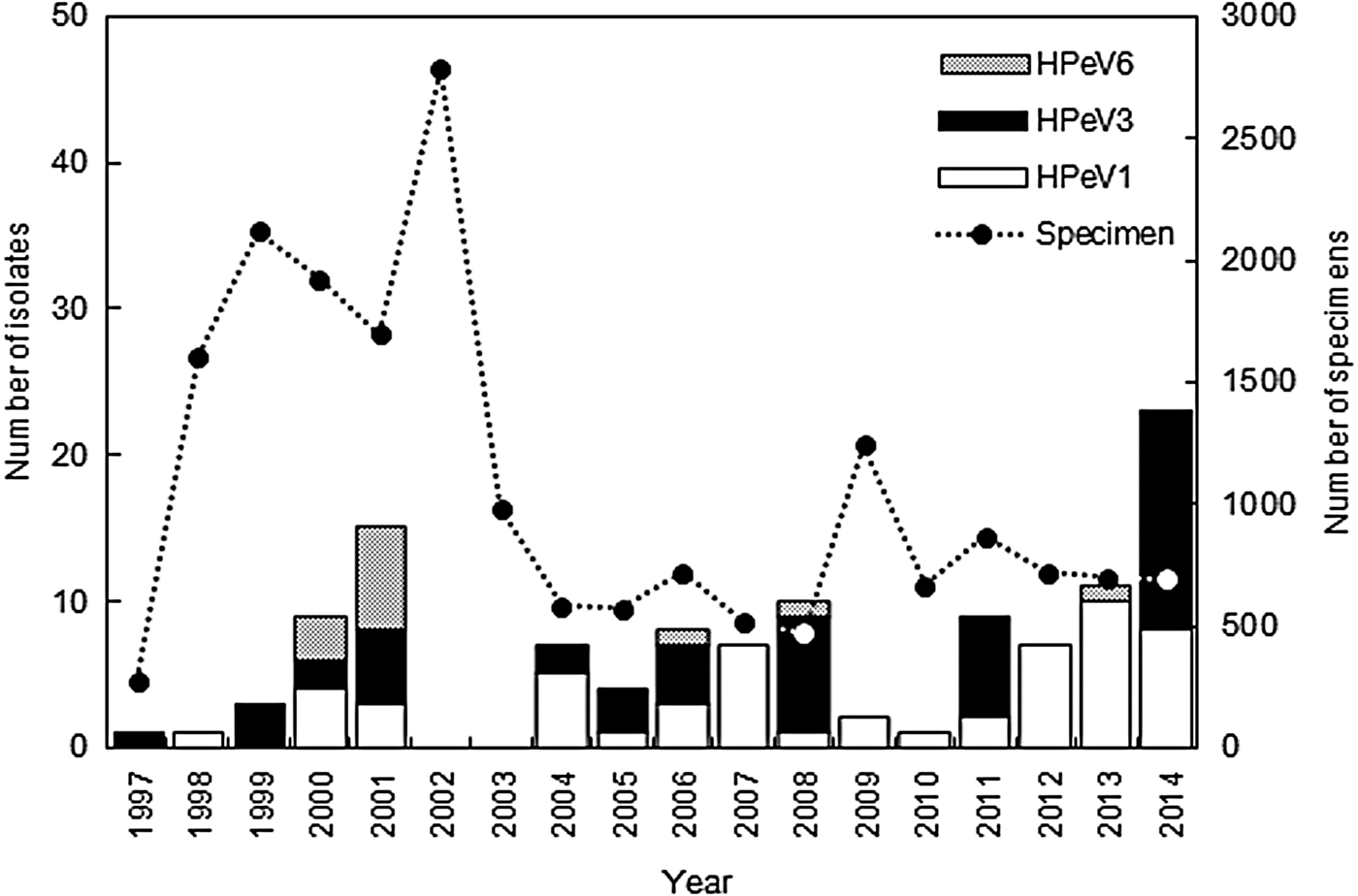
Fig. 1. Yearly distribution and genotype of human parechovirus (HPeV) strains isolated in Niigata, Japan between 1997 and 2014.
HPeV seasonal variation
The numbers of HPeV1 and HPeV3 isolates for each month are summarized in Figure 2a . HPeV1 and HPeV3 were isolated throughout the year, mostly during summer and autumn, followed by winter. They were isolated less during spring. HPeV3 isolates were divided into two groups (Fig. 2b ). One group was isolated in non-epidemic years between 1997 and 2007, and the other group was isolated in epidemic years between 2008 and 2014. Interestingly, HPeV1 and HPeV3 (1997–2007) strains were mainly isolated in August, September and October. HPeV3 (2008–2014) strains were mainly isolated in June, July and August. Two HPeV3 groups showed different seasonal variation by their epidemic pattern. Interestingly, HPeV3 (1997–2007) seasonal variation was very similar to that of HPeV1 (Fig. 2).

Fig. 2. Cumulative number of human parechovirus (HPeV)1 and HPeV3 isolates per month in Niigata, Japan between 1997 and 2014. (a) HPeV1 and HPeV3. (b) HPeV3 (1997–2007) and HPeV3 (2008–2014).
Patients' age distribution
HPeV1, 3, and 6 showed a characteristic distribution by patients' age. Of 118 HPeV isolates, patients' age distribution was between 0 months and 35 years; only one patient was an adult. HPeVs were mostly isolated from patients aged <2 years (95/118, 80·5%); the rest of the paediatric patients were aged 2–9 years. Focusing on genotypes, HPeV1, 3, and 6 were mainly isolated from patients aged 6–11 months (56%), 0–5 months (40%), and ⩾2 years (61%), respectively (Table 2). Although HPeV3 (1997–2007) strains and HPeV3 (2008–2014) strains exhibited different epidemic patterns, the proportion of patients aged 0–5 months was the same at 40% (Table 2).
Table 2. Age distribution and symptoms of patients infected with HPeVs in Niigata, Japan from 1997 to 2014
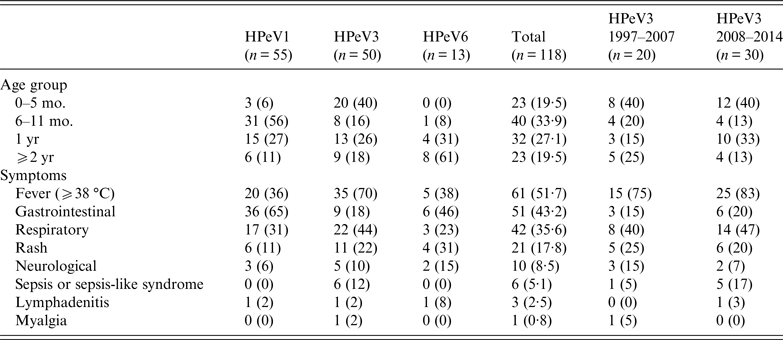
mo., Months; yr, years; HPeVs, human parechoviruses.
Values given are n (%).
Clinical symptoms by HPeV infection
The clinical symptoms were categorized as fever, gastrointestinal symptoms, respiratory symptoms, rash, neurological symptoms (e.g. aseptic meningitis and flaccid paralysis), sepsis or sepsis-like syndrome, lymphadenitis, and myalgia. The most frequent symptom was fever (51·7%) followed by gastrointestinal symptoms (43·2%), respiratory symptoms (35·6%) and rash (17·8%); the frequencies varied between the three genotypes (Table 2). HPeV1 (65%) and HPeV6 (46%) frequently caused gastrointestinal symptoms. Moreover, gastroenteritis patients with HPeV1 and HPeV6 were mainly aged 6 months–1 year and ⩾2 years, respectively. In contrast, HPeV3 most frequently caused fever (70%), and all 20 patients aged 0–5 months developed fever. In addition, only HPeV3 resulted in sepsis or sepsis-like syndrome in patients aged 1 or 2 months, often causing respiratory symptoms. In addition to mild disease, HPeV3 also causes severe disease in neonates and young infants. These results suggested that clinical symptoms were clinically related to HPeV genotype and patients' age.
Genetic differences in genotypes of the HPeV VP1 gene
According to our phylogenetic tree, HPeV1 and HPeV6 were subcategorized into two subgenotypes. HPeV1 and HPeV6 subgenotypes included the prototype strains designated 1A [Reference Benschop25] and 6A, and other strains designated 1B [Reference Benschop25] and 6B, respectively (Fig. 3). All HPeV1 and HPeV6 isolates in Niigata were 1B and 6A, respectively. The pairwise nucleotide and amino-acid sequence identities within HPeV1 were 63–100% and 87–100% and those within the 1B subgenotype were 84–100% and 94–100%, respectively. Similarly, the pairwise nucleotide and amino-acid sequence identities within HPeV6 were 89–100% and 96–100% and those within the 6A subgenotype were 95–100% and 97–100%, respectively. Because the bootstrap value was low in one branch of the HPeV3 phylogenetic tree, HPeV3 was not considered to have subgenotypes. The branch included the HPeV3 prototype strain designated 3A, and all HPeV3 isolates in Niigata were 3A. The pairwise nucleotide and amino-acid sequence identities within the 3A subgenotype were 94–100% and 96–100%, respectively. HPeV1 nucleotide and amino-acid sequence were most divergent in the three genotypes.

Fig. 3. Phylogenetic trees based on nucleotide differences in the capsid protein VP1 [human parechovirus (HPeV)1, 693 bp; HPeV3, 678 bp; HPeV6, 702 bp] of HPeV strains that were mainly isolated in Niigata, Japan between 1997 and 2014. The trees were constructed by the neighbour-joining method with 1000 bootstrap replicates, using MEGA version 6. Bootstrap values >70% are shown. Bars indicate nucleotide divergence. Prototype strains are represented by underlining. The HPeV1 tree includes NII1099–91 which was isolated in 1991 as a reference strain.
Focusing on HPeV3 isolates in 3A subgenotype, there were five lineages. HPeV3 isolates in the epidemic years and in the non-epidemic years formed two (L08/14 and L11) and three lineages (L97/99/00/01, L01/04, and L05/06), respectively. HPeV3 isolates in 2008 and 2014 were closely related genetically, while the isolates in 2014 appeared after an interval of 6 years. Therefore, those isolates formed one lineage (L08/14). HPeV3 isolates in 2011 also formed a lineage (L11) different from L08/14. HPeV3 isolates in the epidemic years showed less diversity than the isolates in the non-epidemic years (Fig. 3).
DISCUSSION
We evaluated seropositivity and epidemiology of HPeVs in Niigata, Japan. The seropositivity to HPeV1, 3, and 6 was higher in older age groups; however, HPeV3 seropositivity was significantly lower in subjects aged >40 years (P < 0·001, P = 0·003). Clinical symptoms of HPeV infection were clinically related to HPeV genotype and patients' age. Moreover, clinical characteristics of HPeV infection were clarified by epidemiology and genotype.
HPeV3 seroprevalence has previously been reported in Japan and Finland. In the Japanese study, serum samples were collected at regional general hospitals, but it was unclear when they were collected. The seroprevalences in the 0–1, 4–6, and >40 years age groups was 15%, 85%, and 87%, respectively [Reference Ito17]. In contrast, in the Finnish study, a large number of serum samples were collected for 14 years from 1994 by the national diabetic birth cohort study; HPeV3 seroprevalences in the 1, 5, and >20 years age groups was <2·7%, <2·7%, and 13%, respectively [Reference Westerhuis18]. The results of the two studies were completely different; the Japanese study had higher levels of neutralizing antibodies to HPeV3. Our results were consistent with those of this previous Japanese study, except that the seroprevalence was significantly reduced in the >40 years age group. In a recent study in Niigata, cord blood samples were collected from September 2013 to January 2014 [Reference Aizawa26]. In an analysis stratified by maternal age, the proportion seropositive for HPeV3 decreased as maternal age increased. Our results were consistent with those reported by this study. These results suggest that people had few opportunities to become infected with HPeV3 (aged >35 years as of 2010). Therefore, HPeV3 activity was probably very low before 1980. Adults may have increased susceptibility to HPeV3 as they lack neutralizing antibodies, which may explain why epidemic myalgia occurs in adults.
In the Finnish study mentioned above, HPeV1 and HPeV6 seroprevalences in the 1, 5, and >20 years age groups were 27%, 83%, and 99% and <12%, 39%, and 57%, respectively [Reference Ito17]. Both HPeV1 and HPeV6 seroprevalences were higher in older age groups and were maintained in adults; however, HPeV6 seroprevalence in each age group was lower than HPeV1 seroprevalence. Our results were consistent with those reported by the Finnish study, with one exception, i.e. Japanese adults had more neutralizing antibodies to HPeV6; however, there were sample year differences, geographical separation, and antigenicity differences in HPeV6.
As per our findings, HPeV1 infection was endemic mainly in gastrointestinal patients aged between 6 months and 1 year, and most children had neutralizing antibodies to HPeV1 by the age of 3 years. HPeV1 seropositivity was maintained in adults. Although HPeV6 infection was sporadically present in circulation, mainly in patients with gastrointestinal symptoms aged ⩾2 years, most children had neutralizing antibodies to HPeV6 by the age of 11–15 years. Maintenance of HPeV6 seropositivity in adults was similar to that of HPeV1 seropositivity. It might be considered that HPeV6 infrequently infects children aged ⩾2 years and, when it does, it results in an asymptomatic infection. In contrast, HPeV3 was mainly isolated from neonates and young infants. Although the proportion of HPeV3 isolation was only 18% at the age of ⩾2 years (Table 2), most children had neutralizing antibodies to HPeV3 by the age of 5 years. Compared to HPeV1, it seems that HPeV3 mostly infects older children and also results in an asymptomatic infection. In the previously mentioned study that was conducted in Niigata, GMTs of antibodies to HPeV1, 3 and 6 showed no significant differences in the three genotypes [Reference Aizawa26]. Our results also show low seropositivity and no significant differences in the 2–6 months age group in the three genotypes (Table 1). Therefore, the question remains that only HPeV3 frequently infects young infants in the three genotypes.
Seasonal variation in HPeVs has been reported [Reference Benschop4, Reference Schuffenecker6, Reference Walters8, Reference Ghanem-Zoubi10–Reference Han11, Reference Ito21] and varies widely with study year, study period, region of the world, and HPeV genotype. In Niigata, HPeV infections also show seasonal variation. HPeV1 strains were isolated less frequently; therefore, HPeV1 isolate data for 18 years between 1997 and 2014 revealed seasonality of HPeV1 infection. Although HPeV3 strains were predominantly isolated in summer, HPeV3 (2008–2014) strains isolated in the epidemic years have had a direct influence on seasonal variation of HPeV3. In contrast, HPeV3 (1997–2007) strains isolated in the non-epidemic years were isolated during summer and winter and were less frequently the same as HPeV1 strains. HPeV3 epidemiology changed not only the yearly distribution but also the seasonal variation.
Phylogenetic trees and nucleotide and amino-acid identities indicated that HPeV1 isolates were the most diverse in the three genotypes. For HPeV3, the isolates in non-epidemic years diverged more than those in the epidemic years. The genetically less diverse lineages circulated epidemically. Therefore, the L08/14, L11 lineage or a new lineage might be expected to recirculate/circulate during the next HPeV3 epidemic. HPeVs yearly distribution was associated with the genetic diversity in HPeV isolates.
This study has some limitations. First, our laboratory examination for HPeV is only virus isolation using a cell culture method; however, the number of HPeVs detected might increase by using RT–PCR or real-time RT–PCR instead of virus isolation. Therefore, the number of HPeV strains might have been underestimated. Second, the HPeV prevalence study did not include a healthy control group, limiting our conclusions with regard to associated symptoms. Third, because serum samples were collected in only a paediatric clinic and from office staff, HPeV seropositivity might not be generalized to other regions of Japan or to other countries. Finally, because the sample numbers of the younger age groups were smaller than the older age groups significant differences in the younger age groups might not be detected.
In conclusion, we clarified the phases of obtaining antibodies to HPeV1, 3, and 6 in children, and adults may have increased susceptibility to HPeV3 as they lack neutralizing antibodies, which may explain why epidemic myalgia occurred in adults. Furthermore, HPeV infection was characterized by the genotype, clinical symptoms, and patients' age. Our findings provide further understanding, especially for milder HPeV infections. Further, we address the question why HPeV3 only frequently infects young infants among the three genotypes by identifying viral receptor and host factors.
ACKNOWLEDGEMENTS
We thank Dr Akihiko Saitoh (Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan) for his critical review and support. We also thank Niigata Prefectural staff for surveillance.
This study received no specific grants from any funding agency, or commercial or not-for-profit sectors.
DECLARATION OF INTEREST
None.








