Overweight and obesity are worldwide public health problems, and are associated with an increase in chronic metabolic diseases such as type 2 diabetes and CVD( Reference Okosun, Liao and Rotimi 1 ). In Mexico, according to a National Nutrition Survey carried out in 2012, the prevalence of overweight and obesity in children (5–11 years old) was 14·46 and 19·8 %, respectively( Reference Gutierrez, Rivera-Dommarco and Shamah-Levy 2 ). According with these findings, Mexico is among the countries with the highest prevalence of childhood overweight and obesity in the world( Reference Gutierrez, Rivera-Dommarco and Shamah-Levy 2 ).
In addition to environmental factors, the genotype explains between 40 and 70 % of BMI variation, mainly by modifying energy expenditure and food intake( Reference Herrera and Lindgren 3 ). The presence of genetic risk factors in obese patients has been associated with a low response to obesity treatments and to an increase in secondary complications associated with this condition( Reference Tounian 4 ). Specifically, the presence of SNP in several genes has been associated with obesity and some of its co-morbidities. Among these genes, the fat mass obesity-associated (FTO), melanocortin 4 receptor (MC4R) and transmembrane protein 18 (TMEM18) have been repeatedly associated with elevated BMI in several populations, both in children and in adults( Reference Lakshman, Elks and Ong 5 , Reference Fawcett and Barroso 6 ). The products of these three genes are expressed in the hypothalamic area, and they have been directly or indirectly related to appetite and energy expenditure regulation( Reference Willer, Speliotes and Loos 7 ). However, the association between genotype and BMI is not always consistent, and it has been observed that the association between the risk allele and the disease varies with ethnicity and populations. In addition, certain effects or associations with metabolic risk factors have been observed depending on the studied age group( Reference Quan, Wang and Tian 8 , Reference Shahid, Rana and Saeed 9 ). The aim of the present study was to investigate a possible association between rs9939609 (FTO), rs17782313 (MC4R) and rs6548238 (TMEM18) SNP and obesity and/or clinical and biochemical alterations in Mexican children.
Methods
Subjects and study design
In this cross-sectional study, a total of 580 school-aged children (8–13 years old) were analysed. Sample size was determined according to the sample size for frequency formula in a population, considering the following parameters: a population size of 25 697 (the number of elementary schools students within the municipality of Queretaro, according to the 2011 census data), a frequency outcome factor in the population of 20 (sd 5) % and a 95 % CI with a design effect of 2·0. The calculated sample size was 488, and a final size of 600 subjects was established to compensate for losses. Proportionate-to-size sampling method was used to select participants in three stages. In stage 1, different schools were chosen by using a stratified random selection considering each district of the municipality as a cluster. In stage 2, within the individual school, two different classrooms were randomly selected, whereas in stage 3 a mean of ten children was randomly selected by the teacher of each classroom. The study was carried out from June 2013 to June 2014. All participants were born in Mexico to parents and grandparents who identified themselves as Mexican-Mestizos. Children with physical and mental abnormalities and with chronic diseases that could alter anthropometry and nutritional or biochemical parameters were not included in the present study.
The study was approved by the Bioethical Committee of the Facultad de Medicina of the Universidad Autónoma de Querétaro (FMUAQ), and was conducted according to the 1964 Helsinki declaration and its later amendments. Signed informed consent was obtained from the parents of all children who participated in this study, after they were fully informed about all the involved procedures. All measurements and samples were collected at the FMUAQ in a single visit.
Anthropometric and physiological measurements
Anthropometry was evaluated by previously trained personnel, following standard procedures( Reference Lohman, Roche and Martorell 10 ). Height was evaluated in the standing position, barefoot and with heels positioned parallel to each other using a stadiometer (Seca® model 240m; Vogel & Halke Gmbh & Co.), with a precision of ±2 mm. To determine fat mass percentage and weight, participants were barefoot with light clothing and were evaluated using a bioimpedance body composition analyser (model SC331S; Tanita Corp. of America Inc.), with a precision of ±0·1 kg. Waist circumference was measured with the children in standing position, at a midpoint between the lower border of the last rib and the upper border of the iliac crest on the horizontal plane, by using an inextensible tape graduated in millimetres.
To determine the nutritional status of children, BMI-for-age Z-scores were calculated and compared with cut-offs and standards from the World Health Organization( 11 ). Children were considered underweight if they had a BMI-for-age Z-score of <−2 sd, normal weight if they had a BMI-for-age Z-score between >−2 and <+1 sd, overweight if they had a BMI-for-age Z-score between >+1 and <+2 sd and obese if they had a BMI-for-age Z-score >+2 sd ( 11 ). Blood pressure was reported as the mean of measurements in both arms with a calibrated paediatric aneroid sphygmomanometer (Riester), according to standard procedures( 12 ). Pre-hypertension and hypertension were considered as a value between the 90th and the 94th percentile and ≥95th percentile, respectively, adjusted for sex, age and height( 12 ).
Biochemical measurements
Fasting blood samples were collected and immediately analysed in an A15 analyzer (Biosystems) for glucose and lipid profile measures (Biosystems). Serum aliquots were obtained and maintained at −20°C for insulin measurement by ELISA (AccuBind ELISA Microwells ISO 13485 & 9001; Monobind Inc.). The homoeostasis model assessment (HOMA-2) was used to evaluate β-cell function (HOMA-B index), insulin sensitivity (HOMA-S index) and insulin resistance (HOMA-IR index). Insulin resistance was considered as a value ≥95th percentile of HOMA-IR index, adjusting for sex and age, using regional reference values for non-obese children( Reference Aradillas-Garcia, Rodriguez-Moran and Garay-Sevilla 13 ). Dyslipidaemias were considered as follows( Reference Peterson and McBride 14 ): hypercholesterolaemia≥5·2 mmol/l, elevated LDL-cholesterol≥3·3 mmol/l, low HDL-cholesterol≤1 mmol/l, and hypertriacylglycerolaemia≥1·1 mmol/l (for ≤9·9 years old) and ≥1·4 mmol/l (for ≥10 years old).
DNA extraction and genotyping
Genomic DNA was extracted from whole, fresh blood by using the Wizard Genomic DNA Purification kit (catalogue no. A1120; Promega). Concentrations of purified DNA samples were spectrophotometrically determined and adjusted to 20–50 ng/ml. Genotyping for rs9939609, rs17782313 and rs6548238 was performed by using TaqMan® SNP Genotyping Assays (catalogue nos C_30090620_10, C_29311887_10 and C_32667060_10 for rs9939609, rs17782313 and rs6548238, respectively; Applied Biosystems). PCR reactions were carried out in a total volume of 25 μl containing 20 ng of genomic DNA, 12·5 μl of TaqMan Universal Master mix (concentration of 2×; Applied Biosystems) and 1·25 μl of TaqMan SNP Genotyping Assay (concentration of 20×; Applied Biosystems) containing both primers and probes. PCR was performed using a StepOne Real time PCR system (Applied Biosystems) under the following conditions: one cycle at 60°C for 30 s, one cycle at 95°C for 10 min followed by forty cycles at 95°C for 15 s and 60°C for 1 min. PCR products were analysed by StepOne™ Software version 2.01 (Applied Biosystems), and alleles were analysed with the default algorithm. Random re-genotyping was conducted in 10 % of the samples to confirm the results.
Statistical analysis
Statistical analyses were performed using Statistical Package for Social Sciences (SPSS) software (version 15; SPSS Inc.). Continuous and categorical variables are displayed as means, standard deviations and percentages, respectively. Differences between categorical variables were analysed by either χ 2 tests or Fisher’s exact tests and the ANOVA for continuous variables. P values<0·05 were considered to be statistically significant. The statistical power for our observed results was calculated by using the formula to calculate power for cross-sectional studies (OpenEpi online software, version 3.01). To do this, a 95 % CI (two-sided) was used, considering exposed subjects as those with FTO and MC4R risk genotypes, non-exposed children as subjects homozygous for the ancestral allele (AA) genotype, and the observed obesity prevalence in each group. Normal and non-normal distributions were determined by the Kolmogorov–Smirnov tests for all variables. Kruskal–Wallis tests were used to assess the differences between medians. One-tailed Spearman’s correlation tests were applied to non-normally distributed data. The Hardy–Weinberg equilibrium was calculated by comparing χ 2 values between the expected and the observed values for genotype counts( Reference Rodriguez, Gaunt and Day 15 ). Linkage disequilibrium was calculated by using Cube X online software( Reference Gaunt, Rodriguez and Day 16 ). Binary logistic regression analyses were performed to associate SNP genotypes with obesity as a dependent variable; age and sex were also included as covariates. ANCOVA were performed in order to evaluate the effect of the SNP, using adiposity as a covariate.
Results
Clinical and biochemical results
A total of 580 children were included with an age of 10 (sd 2·2) years and a female:male ratio of 1:1. Fig. 1 shows a flow diagram of the selection process and the reasons for excluding children and samples. The prevalence of normal weight, overweight and obesity was 59·2, 19·8 and 19·1 %; respectively. General characteristics of the subjects are shown in Table 1. As anticipated, while comparing normal weight children with overweight and obese children, we found significant differences in height-for-age, systolic and diastolic blood pressures, LDL-cholesterol, total cholesterol, TAG, insulin and HOMA-IR, as well as significantly lower values of HDL-cholesterol, HOMA-B and HOMA-S (Table 1).
Fig. 1 Flow chart of the patient inclusion process in the present study.
Table 1 General characteristics observed in the children, according to their nutritional status (Mean values and standard deviations)

Chol, total cholesterol; HOMA-B, homoeostasis model assessment to evaluate β-cell function; HOMA-S, homoeostasis model assessment of insulin sensitivity; HOMA-IR, homoeostasis model assessment of insulin resistance.
a,b,c One-way ANOVA with a Tukey’s post hoc test was used; mean values within a row with unlike superscript letters were significantly different between the values (P<0·05).
* Significantly different values (P<0·05).
† This value includes underweight children.
Insulin resistance was found in 7·30 and 11·47 % among males and females, respectively (9·38 % combined). Pre-hypertension was found in 3·5 and 3·9 % of boys and girls, respectively (3·7 % combined); hypertension was found in 5·83 and 4·6 % of boys and girls, respectively (5·21 % combined). Hypertriacylglycerolaemia was found in 33 %, whereas hypercholesterolaemia was found in 12·8 % of the population. High LDL was found in 3 %, and low HDL levels were found in 25 % of the population.
SNP genotyping
For the three studied SNP, genotypic frequencies were found in the Hardy–Weinberg equilibrium (P>0·05), and genotyping call rates were above 99 % with a re-genotyping concordance rate >99 %. The pair-wise linkage disequilibrium between the three SNP was assessed, and as expected they were found in linkage equilibrium (FTO v. MC4R: D'=0·07, r 2 0·0025, P=0·87; FTO v. TMEM18: D'=0·021, r 2 0·0001, P=1·0; MC4R v. TMEM18: D'=0·06, r 2 0·0033, P=0·99). Table 2 shows allelic and genotypic frequencies for the three SNP.
Table 2 Genotypic and allelic frequencies for r9939609 (FTO), rs17782313 (MC4R) and rs6548238 (TMEM18) and the Hardy–Weinberg equilibrium (HWE)
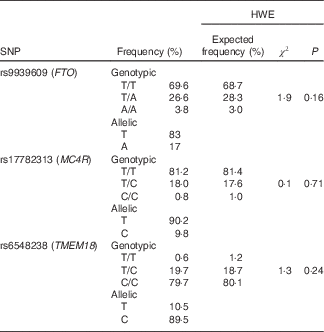
FTO, fat mass obesity-associated; MC4R, melanocortin 4 receptor; TMEM1, transmembrane protein 18.
Association between obesity and genotype
A positive significant association was found between rs9939609 (FTO) homozygous for the risk allele (Hom) and rs17782313 (MC4R) heterozygous (Het) with the presence of obesity (Table 3). There were no obese children with the rs17782313 (MC4R) Hom for the risk allele genotype. The calculated statistical power for our observed results was 97 and 92·3 % for FTO Hom and MC4R Het genotypes, respectively. As for rs6548238 (TMEM18), there was no significant association with obesity. Logistic regression analyses confirmed the association between obesity and genotype for FTO and MC4R risk alleles. In addition, a significant positive association between rs9939609 (FTO) Hom for the risk allele and BMI-for-age Z-score was found. Likewise, there was a significant positive relationship between rs17782313 (MC4R) Het and BMI-for-age Z-score (Fig. 2).
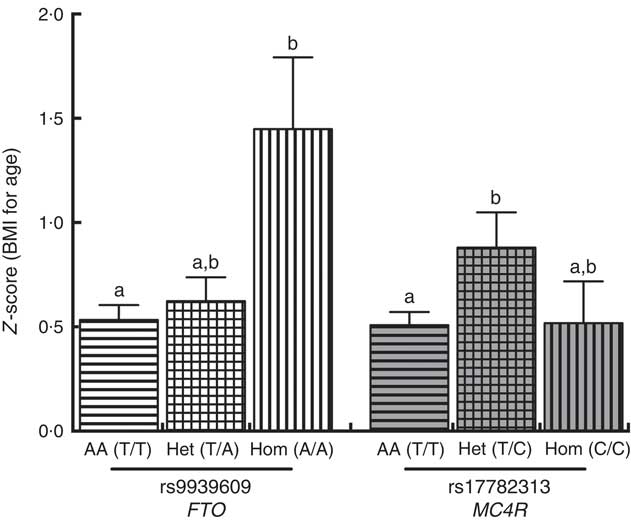
Fig. 2 Comparison between BMI-for-age Z-score and FTO (
![]() ,
,
![]() ,
,
![]() ) and MC4R (
) and MC4R (
![]() ,
,
![]() ,
,
![]() ) genotypes. A Kruskal–Wallis test (Dunn’s multiple comparison test) was used to compare the AA, Het and Hom genotypes. a,b Mean values with unlike letters were significantly different (P<0·05). Values are means, with their standard errors. AA, homozygous for the ancestral allele; Het, heterozygous; Hom, homozygous for the risk allele; FTO, fat mass obesity-associated; MC4R, melanocortin 4 receptor.
) genotypes. A Kruskal–Wallis test (Dunn’s multiple comparison test) was used to compare the AA, Het and Hom genotypes. a,b Mean values with unlike letters were significantly different (P<0·05). Values are means, with their standard errors. AA, homozygous for the ancestral allele; Het, heterozygous; Hom, homozygous for the risk allele; FTO, fat mass obesity-associated; MC4R, melanocortin 4 receptor.
Table 3 Association between obese and normal weight children with the risk genotypeFootnote † (Numbers and percentages; odds ratios, adjusted odds ratios and 95 % confidence intervals)
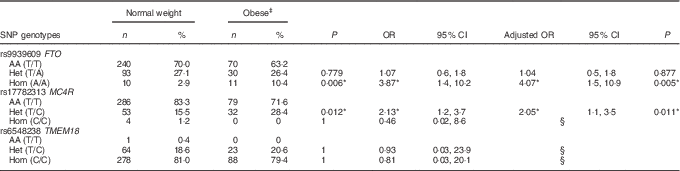
AA, homozygous for the ancestral allele; Het, heterozygous; Hom, homozygous for the risk allele.
* Significantly different values (P<0·05).
† Fisher’s exact test was used for the analysis of this contingency table. The heterozygous and homozygous groups were compared with the ancestral allele group, for the obese v. normal weight children.
‡ There were no obese children with the MC4R (rs17782313) Hom genotype or with the TMEM18 (rs6548238) AA genotype. Binary logistic regressions were used to estimate the adjusted OR by sex and age.
§ Values were not calculated because of the excessive standard error.
Association between genotype and metabolic risk factors
There was no association between hypertensive children and FTO or MC4R genotype. However, while comparing the rs9939609 (FTO) homozygous for the AA in children with the Het group, significantly higher systolic (98·3 (sd 10) v. 102·5 (sd 12) mmHg, P<0·001) and diastolic (67·1 (sd 7) v. 69·3 (sd 8) mmHg, P<0·01) blood pressures were found. In addition, the influence of rs9939609 (FTO) on blood pressure was confirmed by excluding the mediating effect of adiposity by using this variable as a covariate (ANCOVA analysis), with one-tailed P values of 0·0015 and 0·034 for systolic and diastolic blood pressures, respectively.
There was a positive association with the presence of rs17782313 (MC4R) Het genotype and low HDL-cholesterol levels (OR 2·99, P<0·0001; 95 % CI 1·93, 4·64; data not shown). However, when adiposity was used as a covariate (ANCOVA analysis), there was no significant effect of the MC4R genotype. Considering the nutritional status while comparing the AA v. the Het groups, the presence of the rs9939609 (FTO) risk allele was related to a lower fat-free mass percentage (39·2 (sd 9) v. 35·5 (sd 7), P<0·05) and to a higher total cholesterol circulating concentration (4·18 (sd 0·64) v. 4·38 (sd 0·8) mmol/l, P<0·05). Interestingly, these findings were observed only in obese subjects and not in normal weight or overweight children. In addition, while comparing the Pearson’s correlation coefficients between BMI and insulin, total cholesterol and TAG, we found a higher correlation for the Het group compared with AA children, for both FTO and MC4R (Table 4). Nevertheless, only a significant difference between correlation coefficients for BMI and insulin concentration was observed for FTO AA v. the Het children. However, again when adiposity was used as a covariate (ANCOVA analysis), there was no significant effect of FTO genotype on total cholesterol and insulin circulating levels.
Table 4 Comparison of Pearson’s correlation coefficients between BMI and other variables in the presence of fat mass obesity-associated (FTO) and melanocortin 4 receptor (MC4R) alleles of riskFootnote *
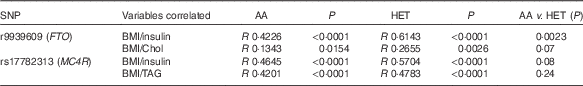
AA, homozygous for the ancestral allele; Het, heterozygous; Chol, total cholesterol.
* Pearson’s correlation test was used to compare both variables, considering a P value <0·05 as significant. To evaluate the difference between the correlation coefficients (R values), Fisher’s transformation (R to Z) was performed.
Discussion
The prevalence of obesity in the population studied was higher compared with the last national prevalence reported in 2012 (19 v. 14·6 %)( Reference Gutierrez, Rivera-Dommarco and Shamah-Levy 2 ). Regarding the rest of the parameters including hypertension, insulin resistance and dyslipidaemias, the population included in the present study was similar to previous reports in open Mexican populations( Reference Romero-Polvo, Denova-Gutiérrez and Rivera-Paredeza 17 – Reference Guerrero-Romero and Rodríguez-Morán 19 ). In addition, and as expected, there were significant differences in most parameters between normal weight and overweight/obese children, including height for age and both systolic and diastolic blood pressures.
It has been reported that the rs9939609 (FTO) risk allele frequency (RAF), the most studied SNP associated with obesity, varies among different populations. In this regard, frequencies above 40 % have been found in Caucasians, whereas frequencies closer to 10 % have been described for Chinese populations( Reference Li, Wu and Loos 20 ). In the present study, rs9939609 (FTO), rs17782313 (MC4R) and rs6548238 (TMEM18) RAF were 17, 9·8 and 89·5 %, respectively. These frequencies are very similar to those reported by León-Mimila et al. ( Reference León-Mimila, Villamil-Ramírez and Villalobos-Comparán 21 ) who found RAF values of 18·1, 8·2 and 91·1 % for rs9939609 (FTO), rs17782313 (MC4R) and rs6548238 (TMEM18), respectively, in Mexican children from Mexico City. Rs17782313 (MC4R) RAF is higher in Caucasian, African-American and Chinese populations (typically >25 %), whereas rs6548238 (TMEM18) RAF is usually >80 % in all studied populations( Reference Loos, Lindgren and Li 22 – Reference Rask-Andersen, Jacobsson and Moschonis 26 ). Therefore, in our population, RAF for FTO and TMEM18 were in the expected range, whereas MC4R was lower compared with internationally reported values.
A positive significant association between the presence of rs9939609 (FTO) and obesity has been consistently described in several populations. A recent meta-analysis showed similar values to data obtained from this study in children and adolescents (OR 1·35; 95 % CI 1·27, 1·44)( Reference Liu, Mou and Cai 27 ) (OR 1·40; 95 % CI 1·15, 1·71)( Reference Quan, Wang and Tian 8 ) and in adults (OR 1·31; 95 % CI 1·26, 1·36)( Reference Peng, Zhu and Xu 28 ). In contrast, León-Mimila et al. ( Reference León-Mimila, Villamil-Ramírez and Villalobos-Comparán 21 ) reported no association between the presence of rs9939609 (FTO) and rs17782313 (MC4R) risk alleles and obesity in Mexican children. Considering that the RAF values were similar, differences regarding genetic background (i.e. interactions with alternative SNP) and/or environmental interactions (i.e. diet and physical activity) could probably explain this discrepancy with our results. However, these authors found an association for this SNP while comparing class III obese v. normal weight adults in a case–control study for this same SNP (OR 1·42; 95 % CI 1·15, 1·76) and for rs17782313 (MC4R) (OR 1·85; 95 % CI 1·23, 2·8). Similarly, Villalobos-Comparán et al. ( Reference Villalobos-Comparán, Flores-Dorantes and Villarreal-Molina 29 ) found a significant association between obesity and the presence of rs9939609 (FTO) in an adult Mexican population, with an OR of 1·38 (95 % CI 1·02, 1·86) and 2·42 (95 % CI 1·71, 3·44) for class I/II and class III obesity, respectively. In accordance with our results, Mejía-Benítez et al. ( Reference Mejía-Benítez, Klünder-Klünder and Yengo 30 ) also found a significant association between obesity and the presence of rs17782313 (MC4R) risk allele (OR 1·4; 95 % CI 1·06, 1·83) while comparing lean v. obese children.
Regarding the association between genotype and metabolic risk factors, Liu et al.( Reference Liu, Mou and Pan 31 ) reported in a recent meta-analysis that the rs9939609 (FTO) risk allele is associated with CVD (OR 1·18; 95 % CI 1·07, 1·30; P=0·001), and this effect seems to be independent of BMI. In the present study, a positive association between rs9939609 (FTO) Het children and higher blood pressure was found after excluding the mediating effect of adiposity. However, we could not find any association with hypertension, which could be explained by the observed low prevalence of hypertension in our sample (5·21 %). Similarly, Pausova et al. ( Reference Pausova, Syme and Abrahamowicz 32 ) reported elevated blood pressure (also independent of adiposity) to be associated with the rs9939609 (FTO) risk allele in a 12–18-year-old French Canadian population. This finding was replicated in an independent group of adults from the same population. He et al. ( Reference He, Fu and Miao 33 ) reported in a recent meta-analysis that Asian obese individuals showed association (OR 1·10; 95 % CI 1·01, 1·19; P=0·032) with the risk of hypertension; remarkably, this association was not observed in non-obese individuals.
Ahmad et al. ( Reference Ahmad, Lee and Pare 34 ) found in a large sample of Caucasian women a higher BMI increase in the presence of the FTO risk allele among those with unhealthy lifestyle (high energy intake and low physical activity). These results suggest that the negative effect of the risk genotype could interact with environmental factors. Although adiposity explained the remaining metabolic alterations, our results also showed an increased strength in the observed relation between BMI and insulin, cholesterol and TAG concentrations in the presence of the risk alleles. Similarly, Villalobos-Comparán et al. ( Reference Villalobos-Comparán, Flores-Dorantes and Villarreal-Molina 29 ) reported a significant association between rs9939609 (FTO) allele of risk and increased cholesterol concentrations among adult obese individuals. It has been described that a healthier lifestyle blunted but did not completely eliminate the associated risk related to the genotype( Reference Ahmad, Lee and Pare 34 ). Taken together, these results suggest that the deleterious effect of the risk genotype is further enhanced in subjects with overweight and obesity.
A major limitation of the present study is the lack of information regarding diet and physical activity of the children. In this regard, it has previously been shown that children with two copies of the lower-risk FTO allele ate less than those with one or two high-risk alleles( Reference Wardle, Llewellyn and Sanderson 35 ). This same study described an association between FTO risk allele with higher consumption of highly palatable food, and this effect was independent of BMI. In addition, children with the FTO risk allele were more likely to have episodes of loss-of-control eating than subjects with the FTO low-risk allele, even after the control of BMI adjusted for age and sex( Reference Tanofsky-Kraff, Han and Anandalingam 36 ). Therefore, this previously obtained data further support our results regarding the observed effect of the genotype in obesity, even in the absence of diet and physical activity data in our sample. In addition, the onset of puberty was not considered in the studied subjects, and this variable also could modify the association between genotype and obesity or metabolic risk factors. This possibility is further supported by the positive association between the risk genotype and the metabolic risk factors observed in adults but not in children.
Conclusion
In the present study, a significant association between obesity and presence of rs9939609 (FTO) and rs17782313 (MC4R) risk alleles was found in Mexican children. In addition, the presence of FTO risk allele was associated with higher blood pressure. Our results also suggest that FTO and MC4R risk genotype could be associated with an increased metabolic risk in children with overweight and obesity, compared with normal weight children. These results suggest that the risk genotype could exert a significant influence on phenotype under the presence of overweight and obesity. Further studies are needed to establish the relationship of the risk genotype with overweight and obesity and the metabolic complications.
Acknowledgements
The authors thank all participants included in the study and their families. The authors also acknowledge Dr Andrés Cruz, Monica Lucio and Alba Nuñez for their technical assistance. The authors acknowledge the support obtained from the Red Nacional para el Tratamiento y Prevención de la Obesidad-CONACYT.
This study was supported by the FOMIX-CONACYT-QRO/2011-C02-175293 granted to J. C. S.-S. and the FOMIX-CONACYT-QRO/2012-C01-192883 granted to P. G.-S.
The author contributions are as follows: P. G.-S., O. P. G. and J. C. S.-S. contributed to the study design and data analyses; M. R.-B., K. F., L. M.-V., C. G.-G., D. G.-G., A. K.-G., O. S.-L. and M. E. V.-H. contributed to subject briefing, data collection and experimental procedures; P. G.-S., M. R.-B., O. P. G., J. L. R., H. L. H.-M. and J. C. S.-S. contributed to interpretation of the findings and writing of the manuscript. All the authors have read and approved the final version of the manuscript.
The authors declare that there are no conflicts of interest.









