Introduction
Rediscoveries of species once considered extinct or extirpated provide rare opportunities for positive messaging about biodiversity and add to a growing list of examples of successful management outcomes for indigenous species affected by non-indigenous invaders (Ladle et al., Reference Ladle, Jepson, Malhado, Jennings and Barua2011; Jones et al., Reference Jones, Holmes, Butchart, Tershy and Kappes2016). Rediscoveries may be particularly likely for secretive taxa that occur in under-surveyed or remote areas (Scheffers et al., Reference Scheffers, Yong, Harris, Giam and Sodhi2011; Caviedes-Solis et al., Reference Caviedes-Solis, Vázquez-Vega, Solano-Zavaleta, Pérez-Ramos, Rovito and Devitt2015), where habitat loss has resulted in scattered populations of a formerly widespread species (Fisher & Blomberg, Reference Fisher and Blomberg2011), where detection probabilities are low to begin with (Durso et al., Reference Durso, Willson and Winne2011; Lee et al., Reference Lee, Fisher, Blomberg and Wintle2017; Thompson et al., Reference Thompson, Koshkina, Burgman, Butchart and Stone2017; Butchart et al., Reference Butchart, Lowe, Martin, Symes, Westrip and Wheatley2018), and where invasive species may be suppressing the densities of individuals below thresholds of detectability but without causing extirpation or extinction (Morrison et al., Reference Morrison, Naikatini, Thomas, Rounds, I. and Thaman Niukula2004).
The 540 km2 Pacific island of Guam is well known for its high proportion of non-indigenous species and the environmental and socio-economic damage they cause (Rodda et al., Reference Rodda, Fritts and Chiszar1997). The ecosystem of this small island is regarded as an example of invasional meltdown, a self-reinforcing mutualism in which the presence of certain non-indigenous species promotes the establishment of other non-indigenous species (Simberloff & Von Holle, Reference Simberloff and Von Holle1999; Christy et al., Reference Christy, Savidge and Rodda2007; Rogers et al., Reference Rogers, Lambers, Miller and Tewksbury2012). Among the numerous non-indigenous animals now present on Guam, some of the most notorious and damaging are the brown treesnake Boiga irregularis, invasive rats Rattus spp., house mouse Mus musculus, marine toad Rhinella marina, musk shrew Suncus murinus, curious skink Carlia ailanpalai, coconut rhinoceros beetle Oryctes rhinoceros, and the little fire ant Wasmannia auropunctata (Fritts & Rodda, Reference Fritts and Rodda1998; Raymundo & Miller, Reference Raymundo and Miller2011; Marshall et al., Reference Marshall, Moore, Vaqalo, Noble and Jackson2017). These and other species have contributed to the dramatic decline, extirpation or extinction of Guam's indigenous birds (Savidge, Reference Savidge1987), lizards (Rodda & Fritts, Reference Rodda and Fritts1992) and mammals (Wiles, Reference Wiles1987).
However, not all species that have been lost from Guam are globally extinct. A small barrier islet known as Dåno′ in the local Chamorro language (colloquially known as Cocos Island) is located 1.6 km off the southern tip of Guam and now serves as both a natural and artificial repository for many taxa that no longer survive on Guam (Fig. 1). Dåno′ is the raised portion (0.39 km2) of the submerged Merizo Barrier Reef (Tracey et al., Reference Tracey, Schlanger, Stark, Doan and May1964). Because of its location on an outer reef, some of the terrestrial invasive species present on Guam have not become established on Dåno′, most notably the brown treesnake. However, brown treesnakes have been sighted, hand captured, or caught in snake traps on Dåno′ since 1988, suggesting that the islet remains vulnerable to introductions (U.S. Geological Survey, unpubl. data.).

Fig. 1 Distribution of the Mariana skink Emoia slevini in the Mariana Islands, showing islands where it is presumed to be extirpated, islands with extant populations, and islands with known extant populations of Emoia atrocostata.
Human land use has drastically altered the habitat on Dåno′ but much of it remains forested (Neubauer & Neubauer, Reference Neubauer, Neubauer and Raulerson1981; McCoid et al., Reference McCoid, Rodda and Fritts1995; Ehrhard, Reference Ehrhard2016). The islet harbours seven habitat types; resort/horticulture, Casuarina forest, mixed strand forest, Pemphis acidula scrub, Scaevola scrub, sand/open areas, and an interior that supports two small wetlands (c. 0.1 ha in total) maintained by subterranean freshwater (Ayers & Vacher, Reference Ayers and Vacher1986). Native species persisting on the island but that are now absent or at low densities on Guam include the Micronesian starling Aplonis opaca, an introduced population of the Guam rail Hypotaenidia owstoni (categorized as endangered under the U.S. Endangered Species Act; U.S. Fish & Wildlife Service, 1984, and Critically Endangered on the IUCN Red List; BirdLife International, 2019), breeding seabirds, and several lizard species that have declined or been extirpated on Guam. The island also supports or has supported several invasive and/or predatory species including Rattus exulans, the monitor lizard Varanus tsukamotoi (recently identified as a species endemic to the Marianas; Weijola et al., Reference Weijola, Vahtera, Lindqvist and Kraus2019), R. marina, C. ailanpalai and possibly M. musculus.
Prior to the introduction of the Guam rail to Dåno′, the Guam Department of Agriculture and the U.S. Department of Agriculture Wildlife Services eradicated R. exulans in 2009 (Lujan et al., Reference Lujan, Vice, Guerrero and Candaso2010). The population of V. tsukamotoi was also culled via trapping and shooting to promote re-establishment of H. owstoni; after 11 months of trapping and shooting in 2011–2012, the estimated density of V. tsukamotoi was reduced from 6.3 lizards per hectare to 1.0 per ha (Ehrhard, Reference Ehrhard2016). Control of monitor lizards has continued at varying levels of intensity since this time.
The Mariana skink Emoia slevini (Åchi ak in Chamorro; Plate 1a) is endemic to the Mariana Islands and is historically known from Guam and Dåno′ (Brown & Falanruw, Reference Brown and Falanruw1972; Brown, Reference Brown1991). It has been documented on 10 islands (from south to north: Dåno′, Guam, Rota, Aguijan, Tinian, Sarigan, Guguan, Alamagan, Pagan and Asuncion), but a recent summary suggested that it persists on only five of these (Sarigan, Guguan, Alamagan, Pagan and Asuncion; USFWS, 2015: Fig. 1). Both the IUCN and U.S. Fish & Wildlife Service list the species as Critically Endangered because of its limited range, recent extirpations on some islands, and presumed declines on islands where it still occurs (Allison et al., Reference Allison, Fisher, Hamilton and Tallowin2013; U.S. Fish & Wildlife Service, 2015). Prior to our study, the last verifiable records for E. slevini on Guam and Dåno′ were from 1945 and 1993, respectively (McCoid et al., Reference McCoid, Rodda and Fritts1995). Dåno′ is also the type locality for E. slevini, and prior to recent molecular work, its closest evolutionary affinities based on colour pattern and size were presumed to be with E. boettgeri and E. arnoensis from the Caroline and Marshall Islands (Brown & Falanruw, Reference Brown and Falanruw1972; Brown, Reference Brown1991).

Plate 1 Representatives of (a) the Mariana skink Emoia slevini and (b) the littoral skink Emoia atrocostata from Dåno′.
In January 2011, c. 21 months after rodent removal and reduction of the monitor lizard population, we observed what appeared to be E. slevini in a small, forested area of Dåno′. Successive visits over the next 2 years resulted in more observations of the species, and in 2014 we non-lethally collected tail tissue from four individuals and used DNA sequence data to verify their identity by comparing results to archived tissue samples of E. slevini collected on Sarigan and Alamagan, located 377 and 653 km north of Guam, respectively. Both islands are part of the Commonwealth of the Northern Mariana Islands and are currently uninhabited (Fig. 1). At the time of our study, these samples represented all known tissues available for DNA sequencing of this rare, endemic species in the Mariana Islands.
Methods
Taxon and field sampling
Emoia slevini is one of 78 currently recognized species of Emoia and a member of the Emoia atrocostata species group (Brown, Reference Brown1991; Plate 1). Richmond et al. (Reference Richmond, Ota, Grismer and Fisher2021) used DNA sequence data to revise the group's membership to include seven nominal taxa: E. adspersa, E. lawesi, E. arnoensis, E. atrocostata, E. boettgeri, E. laobaoense, E. nativitatis and E. slevini (Fig. 2). Emoia adspersa and E. lawesi are endemic to Samoa and nearby islands in Polynesia (Schwaner & Brown, Reference Schwaner and Brown1984) and E. arnoensis and E. boettgeri are endemic to the Marshall and Caroline islands in Micronesia (Brown & Marshall, Reference Brown and Marshall1953). Brown & Falanruw (Reference Brown and Falanruw1972) speculated that E. slevini is most closely related to E. boettgeri, based on morphological similarity and proximity of the Caroline and Marshall Islands to the Marianas. Emoia laobaoense is known from only two specimens collected at a single locality in Viet Nam but has not been observed since its discovery in 1937 (Bourrett, Reference Bourrett1937). Emoia nativitatis is endemic to Christmas Island in the Indian Ocean and was last observed in the wild in late 2009 (Andrew et al., Reference Andrew, Cogger, Driscoll, Flakus, Harlow and Maple2018: Fig. 2). Richmond et al. (Reference Richmond, Ota, Grismer and Fisher2021) identified four regional clades within the wide-ranging E. atrocostata, which they treated as provisional species. One of these is endemic to the Mariana Islands.
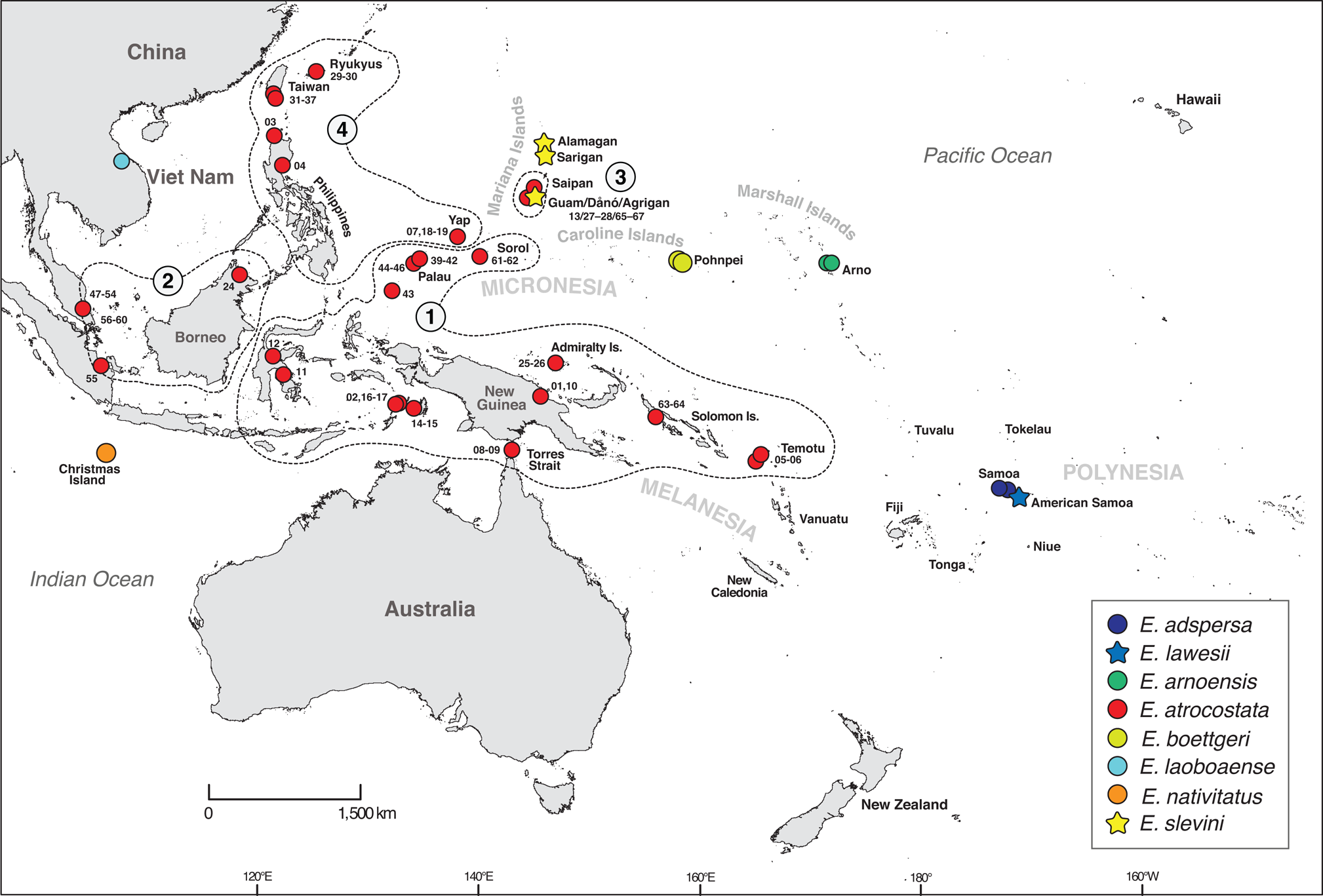
Fig. 2 Sampling map for the phylogenetic analysis of the atrocostata group. Numbered polygons indicate the major regional lineages of E. atrocostata (figure modified from Richmond et al., Reference Richmond, Ota, Grismer and Fisher2021; Fig. 3); numbered sampling sites indicate E. atrocostata included in the phylogenetic analyses (numbers correspond to branches on the mitochondrial genealogy; Fig. 3).
For this study, we added sequence data from the 2014 tissue samples of E. slevini from Dåno′ and Alamagan to the dataset of Richmond et al. (Reference Richmond, Ota, Grismer and Fisher2021), which included sequences from E. slevini previously collected on Sarigan (Smithsonian National Museum of Natural History 536082 and 536083) and from E. atrocostata previously collected on Dåno′ and Guam. The original dataset had multiple representatives of all members of the atrocostata group except for E. laobaoense, and multiple representatives of three outgroup species in the E. cyanogaster group (Brown, Reference Brown1991).
We also added three new tissue samples of E. atrocostata that we collected on Agrigan, a smaller islet c. 7 km east of Dåno′, and three tissue samples from two coastal sites on Saipan, c. 190 km north of Guam. To compare amounts of sequence divergence among all Emoia occurring on Dåno′ and Guam, we sequenced samples of E. cyanura and E. caeruleocauda from Dåno′ and a second sample of E. caeruleocauda from Guam; both are common, widespread species of Emoia and are members of the cyanura species group (Brown, Reference Brown1991). Emoia cyanura is extirpated on Guam, but there is uncertainty as to whether it was native to the Marianas because the only records are from a single locality on Guam and no voucher specimens were ever collected (Rodda et al., Reference Rodda, Fritts and Reichel1991).
We captured all newly collected specimens using adhesive traps placed in shaded areas where we observed lizards foraging (Trapper Max Free, Bell Laboratories, Windsor, USA; see also Rodda et al., Reference Rodda, McCoid and Fritts1993). Lizards were removed from the traps using vegetable oil, which degrades the adhesive, and released on site after taking photographic voucher records. We removed c. 10–15 mm of tail and stored the tissue samples in 95% ethanol until the DNA could be extracted. Locality data for all samples are in Supplementary Table 1.
DNA sequencing
We extracted DNA from tail clips using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germantown, USA) and sequenced the following genes: mitochondrial NADH2 gene (ND2) and adjacent tRNAs (tRNA-trp and -ala) and three nuclear genes: RAG1, Cmos, and R35. We sequenced these genes for consistency with a larger, more comprehensive dataset that includes nearly all species of Emoia (authors, unpubl. data). Primer sequences, PCR reagents, and cycling conditions are detailed in Supplementary Material 1. PCR products were subjected to Sanger dideoxy sequencing using an ABI PRISM 3730 Analyzer (Applied Biosystems, Waltham, USA) at Genewiz, Inc. (La Jolla, USA). We edited the raw sequences using Sequencher 5.0 (Gene Codes Corp., Ann Arbor, USA) and manually aligned them by eye.
Phylogenetic analysis
We first compared the patristic (p) distances among mtDNA haplotypes from separate island populations of E. slevini and among different species in the atrocostata group using DNAsp 6 (Rozas et al., Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017). We did not compare the p-distances based on nuclear alleles because of their high sequence similarity, and instead report only the number of substitutions by which the alleles differed.
To place E. slevini within the broader evolutionary history of the atrocostata group and contextualize the relationships among different island populations, we estimated both a mitochondrial genealogy and the most probable species tree from the set of genealogies inferred for each nuclear and mitochondrial locus (Maddison, Reference Maddison1997; Knowles & Carstens, Reference Knowles and Carstens2007). A detailed description of the phylogenetic methods can be found in Richmond et al. (Reference Richmond, Ota, Grismer and Fisher2021). Data partitions and respective substitution models are provided in Supplementary Table 3. We inferred the mitochondrial genealogy using MrBayes 2.3 (Ronquist et al., Reference Ronquist, Teslenko, Mark, Ayres, Darling and Höhna2012) and estimated divergence times and the species tree using BEAST 2 (Bouckaert et al., Reference Bouckaert, Heled, Kühnert, Vaughan and Wu2014). To calculate divergence times, we allowed the substitution rate to vary independently on each tree branch according to a log-normally distributed rate prior for the ND2 gene (median = 0.0235 substitutions/site/MY, 95% highest posterior density 0.0134–0.0336), which was estimated using an island-age calibration (Richmond et al., Reference Richmond, Ota, Grismer and Fisher2021). The species tree analysis required that the sequences from all samples be assigned to a particular species; we followed Richmond et al. (Reference Richmond, Ota, Grismer and Fisher2021) in treating four regional clades recovered within E. atrocostata as candidate species and considered the remaining nominal species as valid.
Results
Haplotype and allelic diversity
All ND2 sequences of E. slevini from Dåno′ were identical to each other but had unique substitutions compared to two haplotypes from Sarigan, one of which was identical to the single haplotype from Alamagan. These three E. slevini haplotypes differed from each other by only three substitutions and were most similar to haplotypes of E. atrocostata from Agrigan, from which they differed at only six sites. In contrast, haplotypes of E. slevini differ markedly from the two other co-occurring and more ecologically similar species of skink on Dåno′ (E. caeruleocauda and E. cyanura) and Guam (E. caeruleocauda only). Uncorrected pairwise p-distances for ND2 haplotypes collected in the Marianas are shown in Table 1, and a list of haplotypes by name and location are provided in Supplementary Table 2.
Table 1 Uncorrected pairwise p-distances for ND2 haplotypes of Emoia slevini collected in the Mariana Islands. A zero value indicates identical sequences. Species codes are the first two letters of the genus and species, respectivelyFootnote 1. Numerical identifiers following the species code indicate localities (Supplementary Table 1).

1 EMAT, Emoia atrocostata; EMSL, Emoia slevini; EMCY, Emoia cyanura; EMCA, Emoia caeruleocauda.
As expected, based on the limited mtDNA sequence divergence, there was little variation at the three nuclear loci among samples of E. slevini from Dåno′ and Sarigan, and none could distinguish individuals between the two islands. Because of this lack of variation, and added cost, we did not sequence the nuclear loci for the Alamagan samples as mtDNA were sufficient for our purposes. For those samples with nuclear sequence data, we detected (1) three alleles for Cmos, one of which was unique to Sarigan, (2) two alleles for RAG-1, with one unique to Sarigan, and (3) two alleles for R35, both occurring on Dåno′ and only one on Sarigan. The number of polymorphic sites was three, two and two for Cmos, RAG-1 and R35, respectively.
Phylogenetics
The mtDNA genealogy suggests a close relationship between the Dåno′, Sarigan and Alamagan populations of E. slevini and that they diverged from a common ancestor c. 0.27 MYA (range 0.14–0.40; Fig. 3 inset). We also found strong posterior support for a close relationship between E. slevini and the Mariana lineage of E. atrocostata, such that the haplotypes belonging to either species were paraphyletic with respect to each other. Divergence estimates indicate that the two species separated from a common ancestor c. 0.48 MYA (range 0.29–0.68). Together, these two species are the sibling to E. arnoensis, an endemic to the Marshall and Caroline Islands, with all three diverging from a shared ancestor c. 2.78 MYA (range 2.07–3.49; see also Supplementary Fig. 1 for an ultrametric tree with branch lengths scaled according to time).
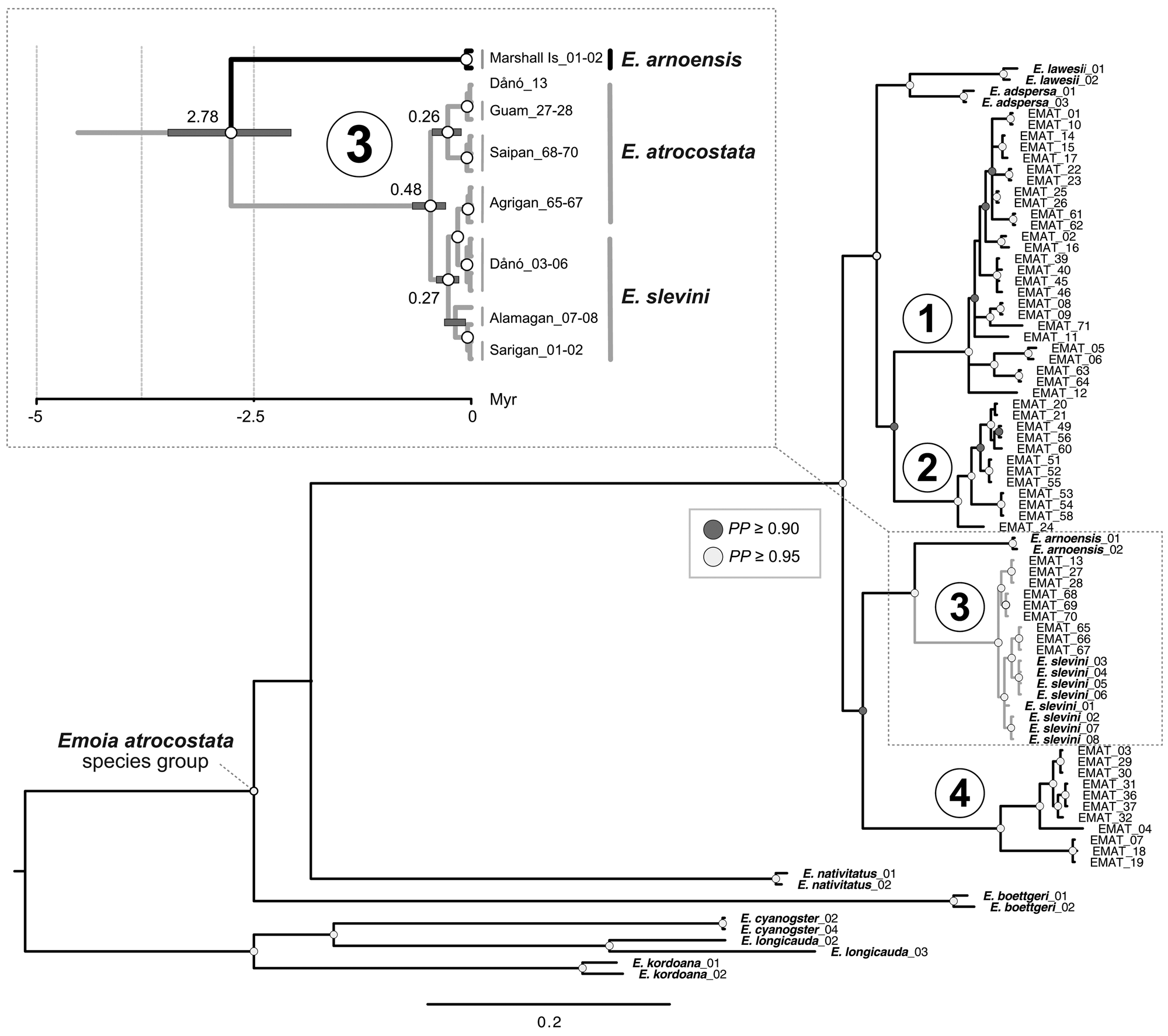
Fig. 3 Mitochondrial genealogy (50% majority consensus tree, branch lengths scaled according to substitutions; PP = posterior probability). The major regional lineages (1–4) of E. atrocostata (EMAT) correspond to the following biogeographical regions: (1) Wallacea, Sahul Shelf, and western Caroline Basin, (2) Sunda Shelf, (3) Mariana Islands, and (4) Philippines, Taiwan, Ryukyu Islands and western Caroline Basin (Fig. 2). Inset shows a close-up of the Mariana lineage, described in the text, with branch lengths scaled according to time in millions of years (grey bars indicate the 95% highest posterior density for the age estimates; numbers on the branches indicate the median of the 95% highest posterior density in millions of years).
Topologies of the nuclear gene genealogies were predictably less resolved than the mtDNA genealogy because of the many fewer phylogenetically informative sites (not shown), with each genealogy showing close relatedness between E. slevini and the Mariana lineage of E. atrocostata. The most probable species tree based on the multispecies coalescent model from this set of genealogies places E. slevini as the sibling to the Mariana lineage of E. atrocostata, with high posterior support (Fig. 4).
Discussion
Samples of the recently detected Emoia from Dåno′ are close genetic matches to E. slevini from Sarigan and Alamagan, confirming they are E. slevini and that the species has apparently persisted at low density on Dåno′ for > 2 decades. The population may be recovering, with individuals now dispersed after being confined to an area of < 300 m2 at the time of the 2011 rediscovery. Rodent eradication appears to be the most likely cause of the recovery of the Mariana skink, but suppression of V. tsukamotoi and/or other as yet unquantified factors may have also contributed. These findings suggest that E. slevini could potentially be surviving on other islands in the northern Marianas where the species is currently considered extirpated or where a lack of records has been accepted as a true gap in its distribution (Rodda et al., Reference Rodda, Fritts and Reichel1991; McCoid et al., Reference McCoid, Rodda and Fritts1995; Reed et al., Reference Reed, Rodda, Siers, Wostl and Yackel Adams2010).
The mitochondrial haplotypes and nuclear alleles belonging to E. slevini and E. atrocostata display a bush-like rather than a tree-like phylogenetic structure as a result of their recent shared ancestry. Speciation is a protracted process such that diverging populations will share a high proportion of the same or similar alleles for extended periods of time until genetic drift and mutations sort the alleles by species (referred to as lineage sorting; Hudson & Coyne, Reference Hudson and Coyne2002; Knowles & Carstens, Reference Knowles and Carstens2007). For slower evolving, bi-parentally inherited nuclear genes, the process is often considerably slower than for rapidly evolving and maternally inherited mitochondrial genes. In these two closely related species, speciation is so recent that the sorting of genetic variation at the targeted loci is incomplete, leading to interdigitating branches in the phylogenetic tree.
Factors determining the distribution of E. slevini
The patchy distribution and decline of E. slevini in the Mariana Islands remain enigmatic, as no obvious pattern exists with respect to underlying causes (McCoid et al., Reference McCoid, Rodda and Fritts1995: Fig. 1). The species has shown a positive response to the removal of invasive ungulates (pigs Sus scrofa and goats Capra hircus) on Sarigan, which has the highest detection rates for E. slevini of any island reported to date (Kessler, Reference Kessler, Veitch and Clout2002, Reference Kessler, Veitch, Clout and Towns2011). Recovery of the species on Sarigan may be primarily a result of forest regeneration following ungulate removal. Our observations on Dåno′ suggest that an even greater response could occur if R. exulans were eradicated from Sarigan.
In New Zealand, negative effects of invasive rodents on the lizard fauna are well documented, with data indicating that severity is dependent on island size, degree of habitat modification, accessibility to refuge habitat, and length of time since the introduction (Whitaker, Reference Whitaker1973; Towns, Reference Towns1991, Reference Towns1994, Reference Towns1996; Newman, Reference Newman1994). In Fiji, endemic iguanas had increased recruitment after eradication of rats and goats (Fisher et al., Reference Fisher, Niukula, Harlow, Rasalato, Chand, Thaman, Veitch, Clout, Martin, Russell and West2019). Predation by introduced rodents also appears to be an important issue for lizard conservation in New Caledonia, as some threatened species have been shown to be regular prey items of the ship rat Rattus rattus and R. exulans (Thibault et al., Reference Thibault, Brescia, Jourdan and Vidal2017).
Emoia slevini and V. tsukamotoi are currently the only endemic species of reptile in the Mariana Islands (Brown & Falanruw, Reference Brown and Falanruw1972; Rodda et al., Reference Rodda, Fritts and Reichel1991; Weijola et al., Reference Weijola, Vahtera, Lindqvist and Kraus2019). However, data from this study and Richmond et al. (Reference Richmond, Ota, Grismer and Fisher2021) suggest that E. cf. atrocostata in the Marianas is a probable third endemic species that merits further attention. Little is known about its distribution and status on other islands (Fig. 1); currently it is known from Dåno′, Guam, Rota, Tinian and Saipan. On Guam, populations persist on small barrier islets (e.g. Agrigan, Fig. 1) at the outer margins of lagoons along the southern edge of the island (Perry et al., Reference Perry, Rodda, Fritts and Sharp1998; Reed et al., Reference Reed, Rodda and Hinkle2007; this study) where they may not be subject to predation by the brown treesnake or other predators.
Other pertinent rediscoveries involving introduced species
Several species of Emoia have been rediscovered on other Pacific islands in recent years, suggesting that at least some can survive at low abundance in the presence of invasive species and/or are capable of finding refugia that insulate them from predation and/or competition. Both phenomena helped explain the 10–fold increase in capture rates for the shore skink Oligosoma smithi in the third year following rat eradications on Korapuki Island, New Zealand, and why other species that coexisted with rats showed changes in both range and productivity once rats were eradicated (Towns, Reference Towns1991).
The introduced big-headed ant Pheidole megacephala was implicated in the decline and presumed extirpation of Emoia impar in the Hawaiian Islands in the early 1900s (Fisher & Ineich, Reference Fisher and Ineich2012). However, in 2000 E. impar was recorded on the offshore islet of Mōkapu, Molokaʻi, where an apparently stable and presumably native population now persists, possibly because the islet lacks P. megacephala (Wood et al., Reference Wood, Burney, Allison and Fisher2013). Similarly, the Fiji barred tree skink Emoia trossula was thought to be extirpated from Vanua Levu, where invasive mammals have been implicated as the primary culprit, but a single individual was observed (and possibly a second one) in 2015 in old growth lowland rain forest (Clause et al., Reference Daugherty, Towns, Atkinson, Gibbs, Towns, Daugherty and Atkinson2018). This suggests that tracts of interior forest may in some cases shelter native species against exposure to invasive rats and mongooses (Olson et al., Reference Olson, Farley, Naisilisili, Raikabula, Prasad, Atherton and Morley2006; Clause et al., Reference Clause, Thomas-Moko, Rasalato and Fisher2018).
Rediscoveries of Emoia in various parts of the Pacific also offer hope for survival of E. atrocostata (described as E. sinus) and the endemic E. nativitatis on Christmas Island in the Indian Ocean (Fig. 2); both are presumed to have recently gone extinct. Smith et al. (Reference Smith, Cogger, Tiernan, Maple, Boland and Napier2012) speculated that increased abundance of the yellow crazy ant Anoplolepis gracilipes and giant centipede Scolopendra subspinipes since the 1990s, combined with the introduction and spread of the Asian wolf snake Lycodon capucinus during a period of unusually dry weather, caused the disappearance of the two Emoia. If these skinks are persisting at low densities, then local eradication of invasive predators may facilitate their recovery. Eradication campaigns targeting the yellow crazy ant A. gracilipes and big-headed ant P. megacephala have had some success, with the highest proportion of successes worldwide occurring for P. megacephala (78% of all documented attempts; see reviews in Hoffman et al., Reference Hoffman, Luque, Bellard, Holmes and Donlan2016). Whether these or other invasive ants pose severe problems in the Marianas is unknown, as the ant communities are poorly characterized for most islands.
Refugia as safeguard against extinction
On biogeographical grounds, a reasonable assumption is that the reptile populations on Dåno′ originated from Guam, because of its proximity and larger size (Rodda et al., Reference Rodda, Fritts and Reichel1991). This displacement of taxa away from Guam and the recent resurfacing of E. slevini are consistent with the range eclipse hypothesis, which predicts that rediscoveries will occur at the edges of a species’ pre-decline range if threats emanate from the interior and force populations into more secure, peripheral habitats (Channell & Lomolino, Reference Channell and Lomolino2000; Hemerik et al., Reference Hemerik, Hengeveld and Lippe2006; Fisher, Reference Fisher2011). A similar pattern exists for New Zealand biota, where losses in biodiversity would be even higher if relict populations were unable to survive on offshore islands that are beyond the dispersal reach of invasive species (Daugherty et al., Reference Daugherty, Towns, Atkinson, Gibbs, Towns, Daugherty and Atkinson1990; Towns, Reference Towns2002).
Dåno′ has become a valuable refuge for endemic biodiversity that either has been lost or is declining in the Marianas, particularly on Guam. There are nine indigenous species of reptiles that historically occurred on both islands (Rodda et al., Reference Rodda, Fritts and Reichel1991); four are now extirpated on Guam, but all persist on Dåno′. In addition to E. slevini, the extirpated species on Guam include E. cyanura, the snake-eyed skink Cryptoblepharus poecilopleurus, and the Micronesia saw-tailed gecko Perochirus ateles (Rodda et al., Reference Rodda, Fritts and Reichel1991). Dåno′ also supports the Micronesian starling Aplonis opaca (rare on Guam but abundant elsewhere in the Marianas) and a reintroduced population of the Critically Endangered flightless Guam rail H. owstoni.
Success in the restoration and enhancement of the Dåno′ ecosystem provides new inspiration for efforts to conserve and restore Guam's biodiversity. It also emphasizes a need for caution in declaring a species extinct or extirpated, as this could detract from surveys that lead to rediscoveries. Alternatively, the rediscovery of a species believed to be extirpated or extinct could spur unsupported optimism for future survival, or perhaps draw the attention of unscrupulous collectors once the rediscoveries are made public (Scheffers et al., Reference Scheffers, Yong, Harris, Giam and Sodhi2011; Meijaard & Nijman, Reference Meijaard and Nijman2014; Caviedes-Solis et al., Reference Caviedes-Solis, Vázquez-Vega, Solano-Zavaleta, Pérez-Ramos, Rovito and Devitt2015). Although the short-term suitability of Dåno′ as a biodiversity refuge has been proven, its long-term status is tenuous. The total island area is 37 ha, with a maximum elevation of 2 m, and only c. 33% of the island is native atoll forest (McCoid, Reference McCoid1996). This small size and low topographic relief make it susceptible to overwash during typhoons. When combined with the persistent threat of non-indigenous species on Guam, these populations of Emoia probably remain at high risk of extirpation. Translocating individuals to managed assurance colonies and/or other offshore islets, parts of Guam, or even other islands in the Marianas merits consideration.
Acknowledgements
We thank L. Berry and J. Liske-Clark for collecting tissue samples of E. slevini from Alamagan; C. Aguon and D. Vice for assistance with scientific collection and export permits; G. Rodda, F. Kraus and an anonymous reviewer for valuable comments on the mansucript; C. Fiedler, B. Lardner, T. Mathies, M. McCoid, N. Sablan and S. Vogt for helpful discussions about E. slevini; S. Hathaway, M. Hogan, K. Kabat, C. Robinson, N. Sablan, T. Tadevoysan and A. Yackel Adams for help with field surveys; and A.J.G. Tornito for providing a translation of the title and abstract into Chamorro (Supplementary Material 2). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author contributions
Study design, fieldwork: JQR, EW, RNR, RNF; laboratory work, analysis: JQR; writing: all authors.
Conflicts of interest
None.
Ethical standards
This study complies with the Oryx guidelines on ethical standards and those outlined in Animal Behaviour (2001) 61, 271–275. Non-lethal tissue sampling was conducted under scientific collecting permits issued to JQR and RNR by the Guam Department of Agriculture/Division of Aquatic and Wildlife Resources and the Commonwealth of the Northern Mariana Islands Division of Fish and Wildlife. Lizards were released at the point of capture and tissue samples from E. slevini were collected prior its listing under the U.S. Endangered Species Act of 1973.









