Levels of circulating fatty acids are associated with risk of incident sudden cardiac arrest (SCA). In particular, higher levels of very long-chain n-3 fatty acids are associated with lower risk and higher levels of trans-fatty acids, especially trans isomers of linoleic acid (trans-18 : 2), are associated with higher risk of SCA( Reference Siscovick, Raghunathan and King 1 – Reference Virtanen, Laukkanen and Mursu 5 ). In addition to circulating fatty acids and other risk factors, common genetic variation may also contribute to SCA risk( Reference Ilkhanoff, Arking and Lemaitre 6 – Reference Sotoodehnia, Siscovick and Vatta 11 ). In agreement with a possible involvement of fatty acid composition in SCA risk, we have provided suggestive evidence that genetic variation in LPCAT1, a gene involved in remodelling of phospholipid fatty acids, is associated with incident SCA( Reference Lemaitre, Johnson and Hesselson 12 ). Whether genetic factors also modify fatty acid associations with SCA is largely unknown.
To investigate possible gene–fatty acid interactions, we used a case-only approach. We measured fatty acids in erythrocyte samples from 1869 SCA patients, on whom we have also assessed genetic variation in fatty acid metabolic pathways( Reference Lemaitre, Johnson and Hesselson 12 ). For this investigation, we focused on genes in metabolic pathways downstream from PUFA, namely eicosanoid pathways, as well as genes involved in the use of fatty acids as an energy source. We hypothesised that variation in these genes would modify the association of circulating DHA, EPA and trans-fatty acids with incident SCA.
Materials and methods
Design
We investigated possible gene–fatty acid interactions on the risk of SCA using a case-only design. Under the assumption of no association in non-cases, coefficients for SNP–fatty acid associations among cases are estimates of coefficients for SNP–fatty acid interactions on the outcome of SCA( Reference Lee, Wang and Cheng 13 ).
Study population
Cases were selected from the Cardiac Arrest Blood Study Repository, a large population-based repository of data and specimens from adult out-of-hospital cardiac arrest patients who were attended by paramedics in Seattle and King county, Washington( Reference Johnson, Lemaitre and Fahrenbruch 14 ). The study was restricted to 1869 SCA patients of European descent with cardiac arrest from 1989 to 2004, with initial rhythm of ventricular fibrillation or asystole, and with fatty acid and genetic data. The research was conducted according to the Declaration of Helsinki. The Human Subject Review Committee of the University of Washington approved the study and the collection of data and specimens in the Cardiac Arrest Blood Study Repository under a waiver of consent.
Blood collection
Paramedics obtained blood specimens from cases in the field after all emergency medical care had been provided and the patient was either clinically stable or deceased, usually within 30 min of the cardiac arrest( Reference Siscovick, Raghunathan and King 1 ). Blood was collected in tubes containing EDTA. Plasma, leucocytes and washed erythrocytes were stored at −80°C. DNA was extracted from leucocytes using standard phenol extraction procedures.
Genotyping and SNP selection
Genotyping was performed on the Affymetrix Axiom panel in the laboratory of P.-Y. K. (Cardiovascular Research Institute, University of California, San Francisco, CA). Sample exclusion criteria were call rates <90 %, sex mismatches or non-European by principal component analysis. SNP exclusion criteria were call rate <97 %, discordant across duplicates or out of Hardy–Weinberg equilibrium (P < 10−5). A set of 522 985 autosomal SNP were imputed into the 566 European Ancestry haplotypes from the 1000G 2010-08 release using minimac( Reference Fuchsberger, Abecasis and Hinds 15 ).
SNP for the study, from candidate gene regions ±50 kB, were extracted from the 1000G 2010-08 release. SNP with a minor allele frequency of <10 % or an imputation quality <0·8 were removed. The selected SNP were intersected with ENCODE regulatory regions from cardiac fibroblasts (HCF), atrial cardiac fibroblasts (HCFaa) or cardiac myocytes (HCM). We used the ENCODE annotation tracks DNAase I hypersensitivity sites, H3K4me3 Histone marks and ChipSeq CTCF sites. All tracks used the HotSpots algorithm for interval selection. Data were extracted from the UCSC Genome Browser (http://genome.ucsc.edu/ENCODE/). SNP falling in at least one ENCODE site were then linkage disequilibrium pruned in PLINK using the genotypes from the European samples of the 2012-03 1000G release and an r 2 threshold of 0·5. A set of 191 SNP in potentially regulatory regions was included in the study. In addition, we examined a SNP in PLA2G2A, rs4654990, that is known to affect circulatory levels of the coded enzyme( Reference Shuvalova, Khasanova and Kaminnaya 16 – Reference Wootton, Drenos and Cooper 18 ).
Fatty acid assays
Fatty acids were extracted using the method of Rose & Oklander( Reference Rose and Oklander 19 ) and fatty acid methyl esters were prepared by direct transesterification using the method of Lepage & Roy( Reference Lepage and Roy 20 ). The fatty acid methyl esters were separated by gas–liquid chromatography using a 100 m × 0·25 mm capillary silica column as described previously( Reference Lemaitre, King and Raghunathan 4 ). A total of forty-two fatty acids were assessed. Fatty acid levels are expressed as percentages of total fatty acids. Trans-isomers with eighteen carbons and one double bond (referred to as trans-18 : 1) are the sum of five isomers; trans-isomers of linoleic acid (trans-18 : 2) are the sum of three isomers.
Statistical methods
Analyses were carried out using Stata 13·0 (StataCorp). Associations of SNP with fatty acid levels were assessed using linear regression. We used additive genetic models adjusted for age, sex and categories of calendar year. Separate models were used for each SNP and each fatty acid. We used a threshold P value for significance of 0·00026 (0·05/191 tested SNP) to correct for multiple comparisons. In sensitivity analyses, we repeated the analyses restricted to SCA patients with initial rhythm of ventricular fibrillation.
Case-only analyses rely on the assumption of no associations in non-cases. To examine this assumption, we looked at the associations of rs4654990 with fatty acids( Reference Mozaffarian, Kabagambe and Johnson 21 , Reference Lemaitre, Tanaka and Tang 22 ) in situ in publicly available results of genome-wide association studies of fatty acids from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (http://www.chargeconsortium.com/main/results).
To obtain estimates of the main effect of fatty acids on SCA risk, we used data from a case–control study that included a subset of the repository cases, without prior diagnosed heart disease, and individually matched controls( Reference Lemaitre, King and Raghunathan 4 ). We used conditional logistic regression to account for matching factors (age, sex and calendar year) and further adjusted for SCA risk factors (smoking, diabetes, hypertension, education, weight, height, leisure time physical activity and fat intake). These analyses, which have been reported earlier( Reference Siscovick, Raghunathan and King 1 , Reference Lemaitre, King and Raghunathan 4 ), were repeated to obtain estimates of the association of one unit higher fatty acid (1 % of total fatty acids) with SCA risk. We did not attempt replication of the case-only findings with the case–control data set because power to detect an interaction of similar magnitude as that of rs4654990 with DHA was only 20 % with the case–control data.
Results
Demographics of the 1869 SCA cases included in the study, and erythrocyte membrane levels of DHA, EPA and trans-fatty acids are shown in Table 1. Levels of DHA correlated with EPA (r 0·57) and levels of trans-18 : 1 with trans-18 : 2 (r 0·47); correlations between DHA/EPA and the trans-fatty acids were negative, ranging from −0·10 to −0·28 (Supplementary Table S1).
Table 1. Characteristics of 1869 sudden cardiac arrest cases in the study
(Mean values and standard deviations, or percentages)
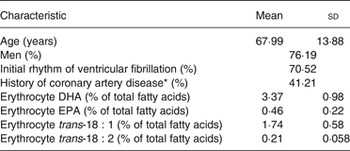
* Among 1206 cases with prior hospital records.
We investigated possible SNP–fatty acid interactions on the risk of SCA in case-only analyses. In the SCA cases, we examined the associations of 191 SNP in regulatory regions of fifty-five candidate genes (Supplementary Table S2) with levels of EPA, DHA and trans-fatty acids. At the preset threshold level of P = 0·00026, no SNP–fatty acid association reached statistical significance (Supplementary Tables S3–S6). The SNP that was most associated with DHA among cases (P = 0·003) was also nominally associated with the other three fatty acids (Table 2). The G allele of rs4654990, near PLA2G2A, with 0·33 allele frequency, was nominally inversely associated with DHA and EPA, and positively associated with trans-18 : 1 and trans-18 : 2 fatty acids. The strongest association was with DHA levels (exponentiated coefficient for one unit (1 % of total fatty acids): 0·905; 95 % CI 0·85, 0·97), indicating that for subjects with a coded allele, the OR associated with one unit higher DHA is about 90 % what it is for subjects with one fewer coded allele.
Table 2. Association of the G allele of rs4654990 with erythrocyte fatty acids in 1869 sudden cardiac arrest cases
(Coefficients with their standard errors)
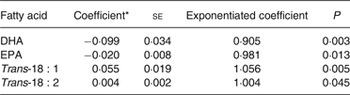
* Coefficient for an increase in one unit of fatty acid (1 % of total fatty acids).
Among the three trans-18 : 2 isomers, both the cis, trans- and the trans, cis-18 : 2 were associated with rs4654990 (regression coefficient: 0·002 (se 0·001) for both isomers; P = 0·048 and 0·041, respectively). In sensitivity analyses restricted to cases found in ventricular fibrillation, we observed similar associations of rs4654990 with fatty acids (Supplementary Table S7).
Coefficients for SNP–fatty acid associations among SCA cases are estimates of coefficients for SNP–fatty acid interactions on the outcome of SCA under the assumption of no association in non-cases( Reference Lee, Wang and Cheng 13 ). We examined this assumption using public data from the CHARGE Consortium( Reference Mozaffarian, Kabagambe and Johnson 21 , Reference Lemaitre, Tanaka and Tang 22 ). In meta-analyses including over 8000 study samples, none of the fatty acids was associated with rs4654990 (P > 0·4; Table 3).
Table 3. Associations of the G allele of rs4654990 with plasma phospholipid levels of fatty acids in meta-analysis results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium*
(Coefficients with their standard errors, and 95 % confidence intervals)
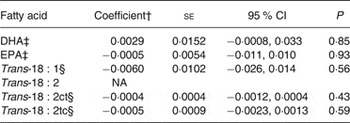
NA, not available.
* Results from http://www.chargeconsortium.com/main/results
† Coefficient for an increase in one unit of fatty acid (1 % of total fatty acids).
‡ Results of associations in 8866 samples( Reference Lemaitre, Tanaka and Tang 22 ).
§ Results of associations in 8013 samples( Reference Mozaffarian, Kabagambe and Johnson 21 ).
Case-only analyses provide estimates of interactions, but not of main effects. A previous report that used the same case population together with population controls showed no main effect of genetic variation in PLA2G2A on SCA risk( Reference Lemaitre, Johnson and Hesselson 12 ). However, we have reported the main effects of fatty acids on SCA risk using a subset of the SCA cases included here and randomly selected, individually matched controls. In particular, we reported associations of higher levels of erythrocyte DHA and EPA with lower risk, and higher levels of trans-fatty acids with higher risk of incident SCA( Reference Siscovick, Raghunathan and King 1 , Reference Lemaitre, King and Raghunathan 4 ). Using the data from that case–control study( Reference Lemaitre, King and Raghunathan 4 ), the estimated regression coefficients for the association of an increase of one unit in fatty acid (1 % of total fatty acids) were −0·32 (se 0·09) for DHA, −0·90 (se 0·41) for EPA, 0·38 (se 0·19) for trans-18 : 1 and 10·32 (se 2·58) for trans-18 : 2. These regression coefficients together with the coefficients from case-only analyses (Table 2), which approximate interaction coefficients, suggest that the associations of fatty acids with SCA risk are more pronounced in the presence of the G allele of rs4654990.
Discussion
In this large case-only study of 1869 SCA cases from the community, we found no evidence for SNP x fatty acid interactions at our preset threshold of statistical significance (P = 0·00026). One SNP near PLA2G2A was nominally associated with higher EPA and DHA levels and lower trans-fatty acid levels among the SCA cases, but not in a large population sample. If replicated, this finding suggests a possible interaction such that n-3 and trans-fatty acids associations with SCA risk may be more pronounced in the presence of the G allele of rs4654990.
The gene PLA2G2A codes for a secretory phospholipase A2 (sPLA2, type IIA). Circulating levels of sPLA2-IIA are associated with incident coronary events and mortality( Reference Kugiyama, Ota and Takazoe 23 – Reference Xin, Chen and Lv 26 ). Whether sPLA2 levels are also associated with SCA risk is not known. Phospholipases A2 release fatty acids from phospholipids, clipping the fatty acid in the sn-2 position where PUFA, including DHA and EPA, are located. The released fatty acid may influence the risk of arrhythmia. For example, in dogs with experimental ischaemia, increased levels of circulating un-esterified DHA protect from ventricular fibrillation( Reference Kang and Leaf 27 ). In addition, PUFA liberated by sPLA2-IIA may be metabolised into eicosanoids( Reference Murakami, Taketomi and Sato 28 ), and prostaglandins derived from EPA and DHA show fewer arrhythmic properties than those from arachidonic acid in cultured myocytes( Reference Li, Kang and Leaf 29 ). Whether the release of DHA or EPA explains an interaction with rs4654990 and enhances these fatty acids’ association with lower SCA risk is not known.
Study strengths include the hypothesis-based investigation, the large number of well-phenotyped incident SCA cases from the community, the use of objective fatty acid biomarkers, and the verification of the case-only assumption in a large dataset from population studies. Several limitations also need to be considered. Blood was collected after cardiac arrest, on average 30 min later; however, n-3 and trans-fatty acid associations with SCA in the study( Reference Siscovick, Raghunathan and King 1 , Reference Lemaitre, King and Raghunathan 4 ) have been replicated in prospective studies( Reference Albert, Campos and Stampfer 2 , Reference Lemaitre, King and Mozaffarian 3 ), suggesting minimal short-term impact on erythrocyte fatty acid composition. We measured fatty acids in different compartments among SCA cases (erythrocyte membranes) and the population sample (plasma phospholipids) used to verify the assumption of no SNP association in non-cases; however, we have shown similar main effects of genetic variants on plasma phospholipid and erythrocyte n-3 and trans-fatty acids( Reference Mozaffarian, Kabagambe and Johnson 21 , Reference Smith, Follis and Nettleton 30 ). Because case-only studies rely on the assumption of no association in non-cases, we were not able to study interactions with other potentially interesting candidate genes, such as the fatty acid desaturase (FADS) genes, that are associated with fatty acids in non-cases( Reference Lemaitre, Tanaka and Tang 22 ). While we selected SNP from regulatory regions, the influence of genetic variation at rs4654990 on PLA2G2A transcription and sPLA2-IIA mass or activity is not known. Another SNP in PLA2G2A that is associated with circulating sPLA2-IIA mass did not show any association with the fatty acids, however, the SNP association with sPLA2-IIA activity appears low and inconsistent( Reference Shuvalova, Khasanova and Kaminnaya 16 ). Due to multiple comparisons, the associations of rs4654990 could be due to chance. In spite of the large number of SCA cases, the study had limited power to detect associations.
In summary, we observed a nominal interaction of a SNP in a phospholipase A2 gene with erythrocyte membrane n-3 and trans-fatty acids, such that the associations of SCA risk with the fatty acids, especially DHA, appear more pronounced in the presence of the G allele of rs4654990. This finding needs to be confirmed.
Supplementary material
The supplementary material for this article can be found at http://www.journals.cambridge.org/10.1017/jns.2016.2
Acknowledgements
The research reported in this article was supported by grants from the National Heart, Lung, and Blood Institute (R01-HL092144, R01-HL092111 and R01-HL088456), the University of Washington Nutrition and Obesity Research Center (P30 DK035816), the Locke Charitable Foundation, the Medic One Foundation, and the Laughlin Family.
D. M. reports ad hoc honoraria or consulting from Bunge, Haas Avocado Board, Nutrition Impact, Amarin, Astra Zeneca, Boston Heart Diagnostics and Life Sciences Research Organization; and scientific advisory board, Unilever North America. The other authors do not report any conflict of interest.
R. N. L. and D. S. S. contributed to the research question, study design, interpretation of the findings and writing of the article; T. M. B., J. A. B. and C. O. J. carried out data analyses; I. B. K., N. S. and T. D. R. contributed to interpretation of the findings and data collection; B. McK. contributed to the study design, interpretation of the findings, and data analyses; D. M. contributed to study design, interpretation of the findings, and writing of the article; S. H. and P.-Y. K. contributed to data collection. All authors read and approved the final version of the manuscript.





