Introduction
Rhigonematids comprise a large and complex group of nematodes with more than 200 species (Hodda Reference Hodda2022) being obligate endoparasites in the posterior gut of millipedes (Arthropoda: Diplopoda) with a monoxenous life cycle (Hunt Reference Hunt1996a). These nematodes are widespread in tropical Africa, but the diversity of rhigonematids from Nigeria is still poorly known, except for Brumptaemilius nigelus and Brumptaemilius sp. (Hunt Reference Hunt2001a, Reference Hunt2001b). Traditionally, species identification of rhigonematid nematodes was mainly based on morphological features, whereas integrative taxonomical approaches, including molecular-based identification, are still scarce (Zhang et al. Reference Zhang, Wang, Hasegawa, Hasegawa, Nagae, Li LW and Li2022). However, some recent studies have provided some ribosomal and mitochondrial (mt) DNA sequence data (i.e., the small subunit ribosomal DNA (18S), the large subunit ribosomal DNA (28S), and the mitochondrial cytochrome c oxidase subunit I (COI) gene, or the mitochondrial genome), which can be used for species identification or phylogeny (Kim et al. Reference Kim, Kim, Cho, Min, Park, Carreno, Nadler and Park2014; Malysheva et al. Reference Malysheva, Mohagan and Spiridonov2015). As a result, the present information on the molecular phylogeny of rhigonematids is far from complete (Mejia-Madrid Reference Mejia-Madrid2018; Zhang et al. Reference Zhang, Wang, Hasegawa, Hasegawa, Nagae, Li LW and Li2022).
Therefore, the main objective of this study was to decipher the biodiversity of this group in the giant millipedes of Nigeria by different ways: i) to anatomically characterise the newly recovered rhigonematids from millipedes in Nigeria by light and scanning electron microscopy; ii) to molecularly characterise these rhigonematids using the D2-D3 28S, ITS and partial 18S rDNA, and cytochrome oxidase c subunit I (COI) gene sequences; and iii) to study the phylogenetic relationships of the identified rhigonematid species with available sequenced species of these nematodes.
Material and methods
Nematode sampling and morphological characterization
During the wet season of 2021, eight samples of millipedes were collected from several localities in southwestern Nigeria (Table 1). Millipede specimens were identified as belonging to the genera Archispirostreptus and Spirostreptus (Table 1). Millipedes were maintained alive in plastic jars until arriving at the laboratory. In the laboratory, millipedes were dissected, with the gut being removed and opened up in a Petri dish containing water to reveal the nematodes therein. Nematodes recovered from the millipede guts were picked out under the stereomicroscope, rinsed in distilled water, and kept in DESS (dimethyl sulphoxide, disodium ethylene diamine tetra-acetic acid, and saturated sodium chloride solution) (Yoder et al. Reference Yoder, De Ley, King, Mundo-Ocampo, Mann, Blaxter, Poiras and De Ley2006) for molecular analyses, whereas those specimens for light microscopy (LM) and morphometric studies were killed by gentle heating in a hot formaldehyde solution and dehydrated using an alcohol-saturated chamber and processed to pure glycerine using Seinhorst’s method (Seinhorst Reference Seinhorst1966) as modified by De Grisse (Reference De Grisse1969) and mounted in permanent slides (De Grisse Reference De Grisse1969). Light micrographs and measurements of the nematodes, including important diagnostic characteristics (i.e., de Man indices, body length, lip region, tail shape, etc.), were done using a Leica DM6 compound microscope with a Leica DFC7000 T digital camera (Leica, Wetzlar, Germany). The raw photographs were edited using Adobe Photoshop v. 22.5.2 (San Francisco, CA, USA).
Table 1. Rhigonematids parasitizing millipedes from Nigeria included in this study

For scanning electron microscopy (SEM), fixed specimens were dehydrated in a graded ethanol-acetone series, critical-point dried, sputter coated with gold according to the protocol by Abolafia et al. (Reference Abolafia, Liébanas and Peña-Santiago2002), and observed with a Zeiss Merlin Scanning Electron Microscope (5 kV; Zeiss, Oberkochen, Germany).
Specimens for molecular analysis were stored in 2-ml Eppendorf tubes containing DESS and sent from Nigeria to the Institute for Sustainable Agriculture, Córdoba, Spain.
Molecular characterization and phylogenetic analyses
For molecular analyses, single rhigonematid specimens were temporarily mounted in a drop of 1M NaCl containing glass beads (to avoid nematode crushing/damaging) to ensure that specimens conformed with the target species. All necessary morphological and morphometric data were recorded. This was followed by DNA extraction from single individuals as described by Archidona-Yuste et al. (Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016). DNA extraction was always based on single nematode specimens and, more decisively and without exception, all the molecular markers came from the same single DNA extracted nematode in individual PCR tubes for all the rhigonematids species. The D2-D3 segments were amplified using the D2A (5’-ACAAGTACCGTGAGGGAAAGTTG-3’) and D3B (5’-TCGGAAGGAACCAGCTACTA-3’) primers (De Ley et al. Reference De Ley, Félix, Frisse, Nadler, Sternberg and Thomas1999). The Internal Transcribed Spacer region (ITS) was amplified using forward primer 18S (5’-TTGATTACGTCCCTGCCCTTT-3’) and reverse primer vrain2r (5’-TTTCACTCGCCGTTACTAAGGGAATC-3’) (Vrain et al. Reference Vrain, Wakarchuk, Lévesque and Hamilton1992). The portion of 18S rRNA was amplified using primers 988F (5’-CTCAAAGATTAAGCCATGC-3’), 1912R (5′-TTTACGGTCAGAACTAGGG-3’), 1813F (5’-CTGCGTGAGAGGTGAAAT-3′), and 2646R (5′-GCTACCTTGTTACGACTTTT-3′) (Holterman et al. Reference Holterman, Van Der Wurff, den Elsen, van Megen, Bongers, Holovachov, Bakker and Helder2006). Finally, the partial COI gene was amplified by PCR using the forward primer LCO (5′-GGT CAA CAA ATC ATA AAGATA TTG G-3′) and the reverse primer HCO (5′-TAAACT TCA GGG TGA CCA AAA AAT CA-3′) (Black et al. Reference Black, Hoeh, Lutz and Vrijenhoek1994).
All PCR assays were done according to the conditions described by Archidona-Yuste et al. (Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016). The amplified PCR products were purified using ExoSAP-IT (Affimetrix, USB products) and used for direct sequencing on a DNA multicapillary sequencer (Model 3130XL genetic analyser; Applied Biosystems, Foster City, CA, USA), using the BigDye Terminator Sequencing Kit V.3.1 (Applied Biosystems, Foster City, CA, USA) at the StabVida sequencing facilities (Caparica, Portugal). The newly obtained sequences were submitted to the GenBank database under the accession numbers indicated on the phylogenetic trees.
Phylogenetic analyses
D2–D3 expansion segments of 28S, ITS and 18S rDNA, and COI mtDNA sequences of the recovered rhigonematid species were obtained in this study. These sequences, and other sequences from species of the Suborder Spirurina from GenBank, were used for phylogenetic analyses. Outgroup taxa for each dataset were chosen following previously published studies (Malysheva et al. Reference Malysheva, Van Luc and Spiridonov2012; Kim et al. Reference Kim, Kim, Cho, Min, Park, Carreno, Nadler and Park2014; Malysheva et al. Reference Malysheva, Mohagan and Spiridonov2015; Mejia-Madrid Reference Mejia-Madrid2018; Zhang et al. Reference Zhang, Wang, Hasegawa, Hasegawa, Nagae, Li LW and Li2022). Multiple sequence alignments of the different genes were made using the FFT-NS-2 algorithm of MAFFT V.7.450 (Katoh et al. Reference Katoh, Rozewicki and Yamada2019). Alignments were manually edited and trimmed of the poorly aligned positions (Hall Reference Hall1999) using a light filtering strategy (up to 20% of alignment positions), which has little impact on tree accuracy and may save some computation time, as suggested by Tan et al. (Reference Tan, Muffato, Ledergerber, Herrero, Goldman, Gil and Dessimoz2015). Phylogenetic analyses of the sequence datasets were based on Bayesian inference (BI) using MrBayes 3.1.2 (Ronquist & Huelsenbeck Reference Ronquist and Huelsenbeck2003). The best-fit model of DNA evolution was obtained using JModelTest V.2.1.7 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012) with the Akaike information criterion (AIC). The best-fit model, the base frequency, the proportion of invariable sites, the gamma distribution shape parameters, and substitution rates in the AIC were then used in MrBayes for the phylogenetic analyses. BI analyses were performed under a general time-reversible model with invariable sites and a gamma-shaped distribution (GTR + I + G) for the D2–D3 segments of 28S and ITS rDNA, SYM model with invariable sites and a gamma-shaped distribution (SYM + I + G) for the partial 18S rDNA, and a transversion model with invariable sites and a gamma-shaped distribution (TVM + I + G) for the partial COI gene. All Bayesian analyses were run separately per data set with four chains for 4 × 106 generations. The Markov chains were sampled at intervals of 100 generations. Two runs were conducted for each analysis. After discarding burn-in samples of 30% and evaluating convergence, the remaining samples were retained for in-depth analyses. The topologies were used to generate a 50% majority-rule consensus tree. Posterior probabilities (PP) were given on appropriate clades. Trees from all analyses were visualised using FigTree software version v.1.42 (Rambaut Reference Rambaut2014). A combined analysis of the three ribosomal genes was not undertaken due to some sequences not being available for all species.
Results
Low to high densities of the presently studied rhigonematids were detected in the intestines of species of black African and brown giant millipedes of the genera Archispirostreptus and Spirostreptus from Nigeria. Detailed morphological, morphometrical, and molecular information for these species is provided below, confirming its identity. However, only one single unidentified juvenile specimen was sequenced for ribosomal and mitochondrial markers but cannot be assigned to any genera or species (Table 1).
Taxonomy
Phylum: Nematoda Rudolphi, 1808
Class: Chromadorea Inglis, 1983
Order: Rhigonematida Inglis, 1983
Suborder: Rhigonematina Inglis, 1983
Superfamily: Rhigonematoidea Artigas, 1930
Family: Rhigonematidae Artigas, 1930
Genus: Rhigonema Cobb, 1898
Family: Xustrostomatidae Hunt, Reference Hunt2002
Genus: Obainia Adamson, Reference Adamson1983
Superfamily: Ransomnematoidea Travassos, 1930
Family: Carnoyidae Filipjev, 1934 (Travassos & Kloss 1960)
Genus: Brumptaemilius Dollfus, 1952
Genus: Gilsonema Hunt, Reference Hunt1998
Brumptaemilius sp
Description
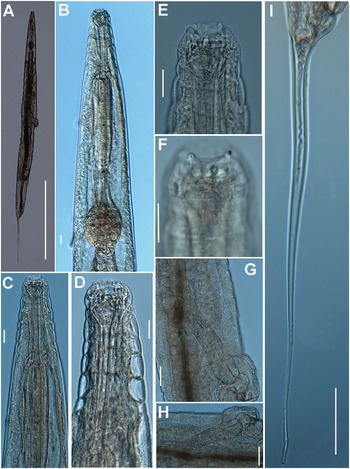
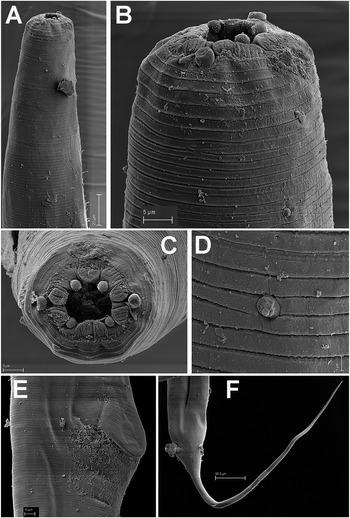
Female. Body small to medium size, straight after heat relaxation, with a long filiform tail (Figure 1A,I). Cuticle with fine transverse striae. Lip region low and rounded, oral disc with four, rounded cephalic papillae (Figure 2B,C). Amphidial apertures prominent, located at junction of oral disc and subsequent annulus. Peg-like somatic papillae present along whole body. Pharynx typical of the genus, with stoma expanding abruptly into a cylindrical corpus (189, 191 μm long) and posteriorly offset from a narrow isthmus (70, 74 μm long). Basal bulb subspheroid (73, 87 μm wide). Vulva about halfway down the body (Table 2), covered by a conspicuous vulval flap 122, 126 μm long. Ovejector structure conforming to Type 1 (Adamson Reference Adamson1987), (374, 386 μm long). Two ovaries reflexed, often containing ovoid eggs, measuring (140–68 μm). Tail large size, filiform.

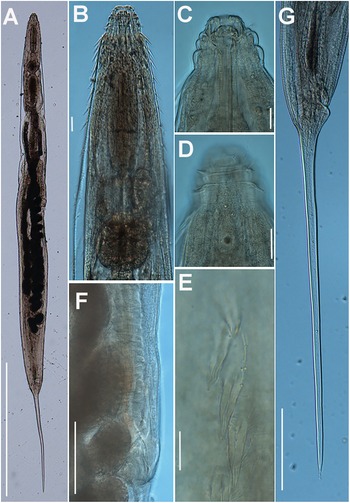
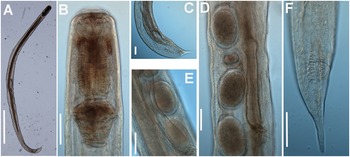
Figure 1. Brumptaemilius sp. LM micrographs of female. A: Entire body; B: Pharyngeal region; C–F: Detail of anterior region; G, H: Detail of vulval region; I: Female tail. (Scale bars: A = 1000 μm; B–F = 20 μm; G, H = 50 μm; I = 100 μm).
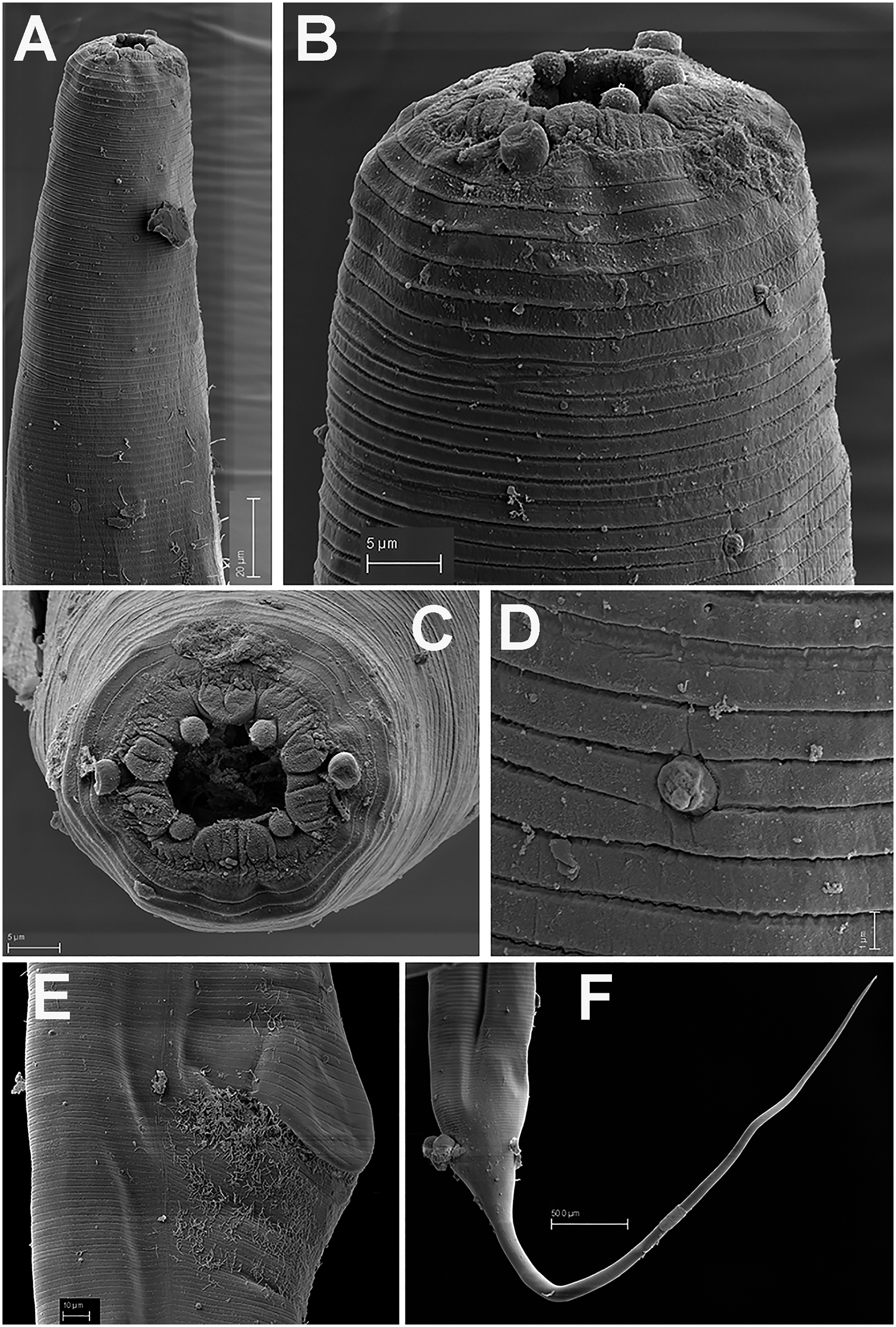
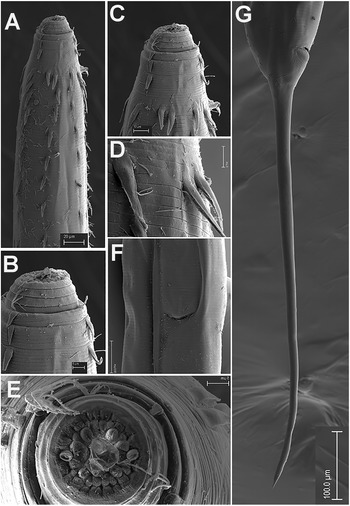
Figure 2. Brumptaemilius sp. SEM micrographs of female. A, B: Lip region; C: En face view; D: Detail of a body papilla; E: Detail of vulval region showing anterior vulval flap; F: Female tail. (Scale bars: A = 20 μm; B–C= 5 μm; D = 1 μm; E = 10 μm; F = 50 μm).
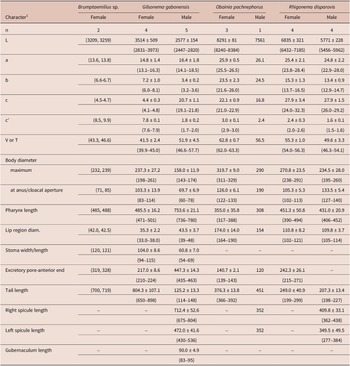
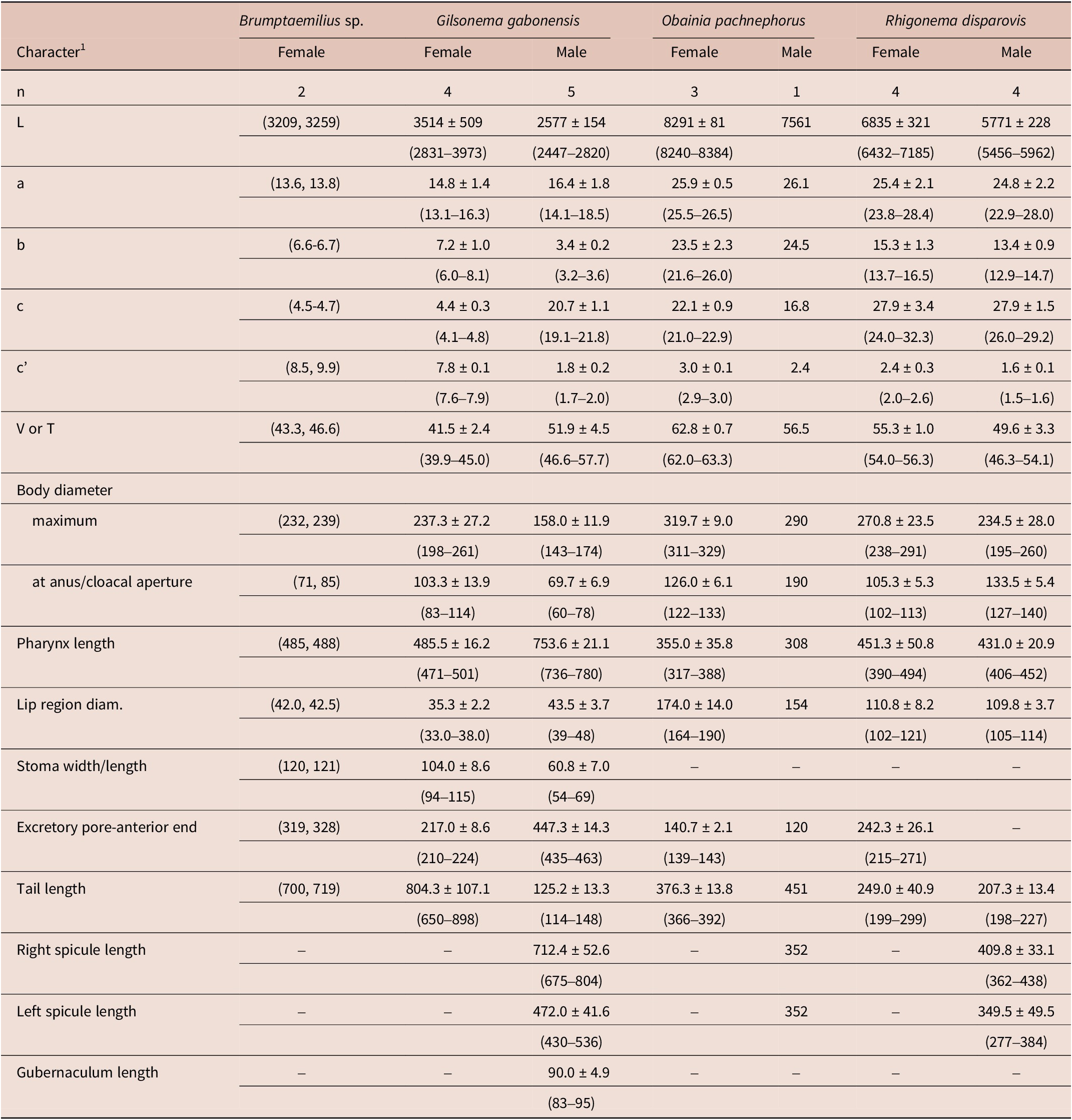
Table 2. Morphometrics of rhigonematids from Nigeria. All measurements are in μm and in the form: mean ± s.d. (range)
Remarks
Morphology and morphometrics of the Nigerian specimens (Table 2) agree well with morphological features of the genus Brumptaemilius. However, although several juveniles were detected and sequenced, these specimens could not be matched to any species due to the absence of the male (despite several trials in millipede extraction), which calls for the need for additional studies in this and/or other areas of Nigeria.
Gilsonema gabonensis (Adamson Reference Adamson1983) Hunt Reference Hunt1998
Description
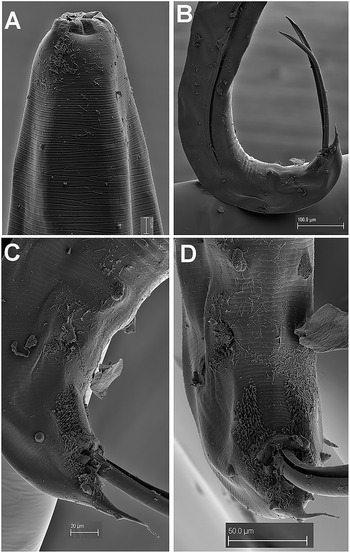
Male. Body small to medium size, cylindrical, slightly tapering and with greater curvature in posterior end (Figure 3). Cuticle finely annulated transversally, lip region conoid-rounded (44 ± 3.7 (39–48) μm wide) (Figure 4A). Pharynx narrowed anteriorly, forming a collar around posterior stoma and then soon expanding into a cylindrical corpus (532 ± 36.7 (507–594) μm long) posteriorly offset from a narrow isthmus. Basal bulb well developed, subspheroid (85 ± 7.5 (75–96) μm wide). Nerve ring located in the posterior part of corpus, (517 ± 14.0 (503–531)) μm from anterior end. Excretory pore located about middle of corpus. Testis beginning in posterior half of body, flexing posteriorly and leading to seminal vesicle and glandular vas deferens. Tail short, conoid, with a terminal projection. Bursa absent. Spicules very long, ventrally arcuate, almost isometric and isomorphic with transverse striae (Figures 3,4). Gubernaculum prominent, with dorsal hole. Eight copulatory papillae present, three pre-cloacal pairs and two pairs postcloacal and one unpaired dorsal papilla at the beginning of the terminal tail projection (Figure 4B–D). Area rugosa consisting of precloacal and postcloacal fields of small round bosses; lateral to these, striae modified with small swellings, and several irregular rows of microtrich formations (Figure 4B–D).

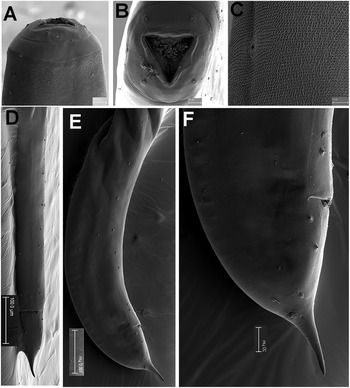
Figure 3. Gilsonema gabonensis (Adamson Reference Adamson1983) Hunt Reference Hunt1998. LM micrographs of male. A: Entire body; B: Pharyngeal region; C–E: Detail of anterior region; F: Male tail with detailed position of post-cloacal papillae. (Scale bars: A = 200 μm; B–F = 50 μm; G = 20 μm; H = 20 μm).
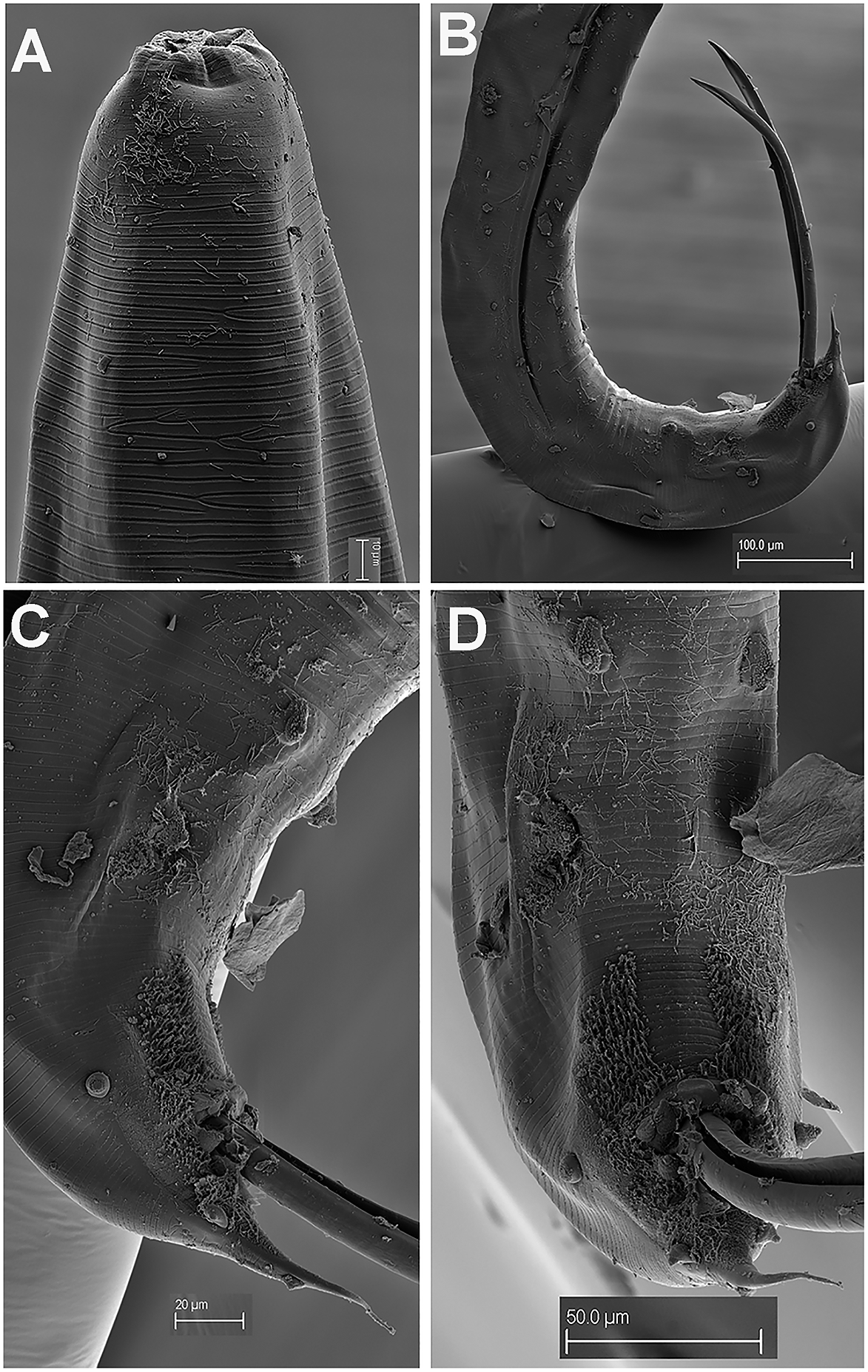
Figure 4. Gilsonema gabonensis (Adamson Reference Adamson1983) Hunt Reference Hunt1998. SEM micrographs of male. A: Lip region; B: Tail region; C–D: Detail of male tail showing pre- and post-cloacal papillae. (Scale bars: A = 10 μm; B = 100 μm; C = 20 μm; D = 50 μm).
Female. Body stout medium size with a long filiform tail (Figure 5A). Lip region round, with 3–4 prominent transverse annuli (Figure 6B–D). Cuticle with fine transverse striae bearing longitudinal rows of protuberant cuticular spines (24 ± 2.4 (21–26) μm long) in the anterior region (Figure 5B–E; Figure 6A–D). Oral aperture circular under SEM, 20–22 μm wide (Figure 6E). Pharynx narrowing anteriorly, forming collar around posterior stoma and then expanding abruptly into a relatively long, narrow, cylindrical corpus (218 ± 15.9 (200–238) μm long) posteriorly offset from a narrow isthmus (76 ± 8.0 (64–81) μm long). Basal bulb subspheroid (96.8 ± 7.4 (89–104) μm wide). Nerve ring crossing just before cylindrical part of corpus (328 ± 12.7 (319–337) μm from anterior end). Excretory pore usually located within posterior third of cylindroid part of corpus. Vulva slightly anterior to median. Anterior lip distended by the presence of a massive, amber-brown copulatory plug lodged under and around the lip. Vagina long, muscular, anteriorly directed before reflexing to common uterus. Two ovaries present, running anteriorly. Ovejector structure conforming to Type 1 (Adamson Reference Adamson1987), 288 ± 76.1 (183–359) μm long. Tail filiform, large size, tapering steadily to terminus (Figure 5G, Figure 6G).

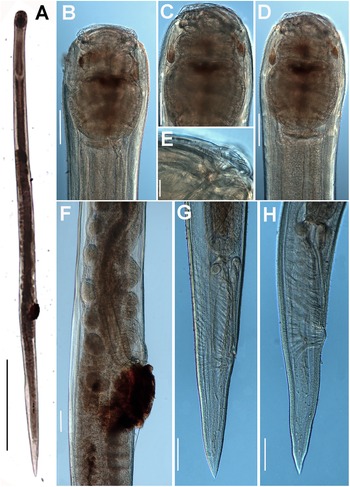
Figure 5. Gilsonema gabonensis (Adamson Reference Adamson1983) Hunt Reference Hunt1998. LM micrographs of female. A: Entire body; B: Pharyngeal region; C–D: Detail of anterior region; E: Detail of cervical spines; F: Detail of vulval region; G: Female tail. (Scale bars: A = 1000 μm; B–D = 20 μm; E = 10 μm; F = 100 μm; G = 100 μm)

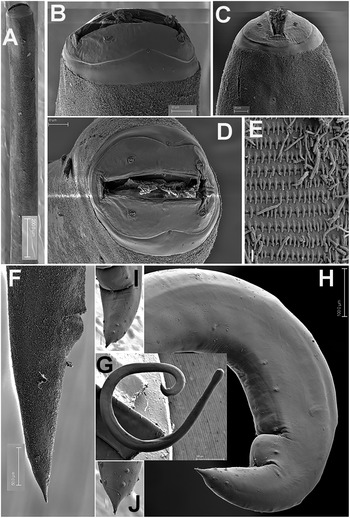
Figure 6. Gilsonema gabonensis (Adamson Reference Adamson1983) Hunt Reference Hunt1998. SEM micrographs of female. A–C: Anterior region showing spines; D: Detail of spines; E: En face view; F: Detail of vulval region; G: Female tail. (Scale bars: A = 20 μm; B = 5 μm; C = 10 μm; D = 2 μm; E = 5 μm; F = 50 μm; G = 100 μm).
Remarks
Morphology and morphometrics of the Nigerian specimens (Table 2) agree with those of the type population of this species from Gabon (Adamson Reference Adamson1983). The main differences between the Nigerian population of G. gabonensis and the original population of this species are a slightly thinner body (198–261 vs 278–301) μm, a slightly shorter corpus length (200–238 vs 188–301) μm, and a slightly shorter tail (650–898 vs 905–1054) μm. The small morphometric differences detected may be due to geographical intraspecific variability. To our knowledge, this is the first report of this species from Nigeria.
Obainia pachnephorus Hunt Reference Hunt1996b
Description
Male. Body cylindrical slightly tapering and with greater curvature in posterior end (Figure 7). Lip region laterally compressed; oral aperture a dorso-ventrally elongate slit (Figure 8A–D). Cuticle with fine transverse striae bearing delicate microtrichs (1.–-2.0 μm long) in the anterior region (Figure 8A–E), becoming shorter thereafter (ca 1 μm). Elongate labial cap in en face view (174 ± 14.0 (164–190) μm wide), with four equidistant prominent papillae (two sub-dorsal and two sub-ventral) (Figure 8D). Oral aperture elongate-oval under SEM, 100–135 μm wide (Figure 8B–D). Pharynx short and powerful, with a short and stout corpus (131 ± 4.0 (127–135) μm long). Isthmus absent, basal bulb well-developed as long as wide (221 ± 8.6 (212–229) μm wide) occupying more than 50% of pharynx. Nerve ring located just anterior to basal bulb (108 ± 4.5 (103–112) μm from anterior end). Tail medium size, conoid, ending in a digitiform process. Bursa absent. Spicules ventrally arcuate, almost isometric and isomorphic. Twenty-three copulatory papillae present arranged as four pre-cloacal pairs, a single mid-ventral papilla on anterior cloacal lip, and seven pairs postcloacal (Figure 8G–J).
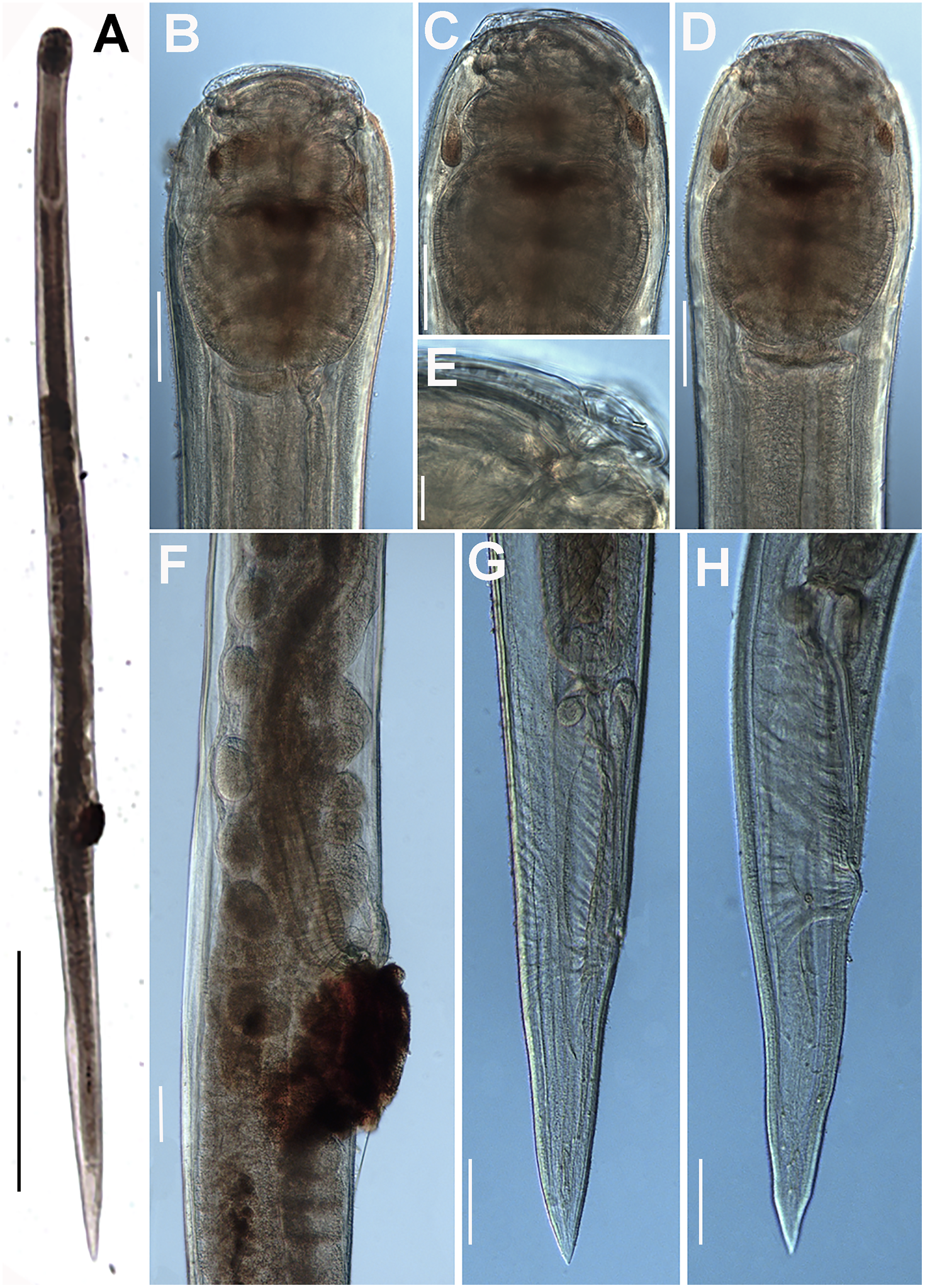
Figure 7. Obainia pachnephorus Hunt, Reference Hunt1996b. LM micrographs of female. A: Entire body; B–D: Lip region; E: Detail of labial papilla; F: Detail of vulval region; G, H: Female tail. (Scale bars: A–D = 100 μm; E = 10 μm; F–H = 100 μm).

Figure 8. Obainia pachnephorus Hunt, Reference Hunt1996b. SEM micrographs of female and male. A: Female anterior region; B–D: Detail of female lip region; E: Detail of microtrichs; F: Female tail; G: Entire male; H: Male tail region; I, J: Detail of male tail showing papillae. (Scale bars: A = μm; B–D and I, J = 20 μm; E = 2 μm; F = 50 μm; G = 500 μm; H = 100 μm)
Female. Body slightly larger than male. Morphology almost identical to that of male, except for sexual characters. Vulva post-median (Table 2), covered by a conspicuous vulval flap with brown deposits around it. Vagina long. Ovejector structure conforming to Type 2 (Adamson Reference Adamson1987), 678 ± 131.6 (595–830) μm long. Ovaries reflexed, often containing ovoid eggs with smooth shell, measuring 100 (93–111) x 65 (63–68) μm (Figure 7). Tail medium size, conoid, tapering steadily to terminus.
Remarks
Morphology and morphometrics of the Nigerian specimens (Table 2) agree with those of the type population of this species from Ivory Coast (Hunt Reference Hunt1996b). The main differences between the Nigerian population of O. pachnephorus and the original population of this species are based on a single male specimen detected in our study: a slightly longer body (7561 vs (5190–5780) μm); slightly wider maximum body diameter (290 vs (198–226) μm); and spicule length (352 vs (280–367) μm). These small morphometrics differences may be due to geographical intraspecific variability. To our knowledge, this is the first report of this species from Nigeria.
Rhigonema disparovis Van Waerebeke Reference Van Waerebeke 1991
Description
Male. Body cylindrical gradually tapering and with greater curvature in posterior end (Figure 9). Cuticle apparently smooth, covered with numerous fine and short microtrichs (1.5–2.0 μm long) in the anterior region (Figure 10A, C). Labial cap circular in en face view (110 ± 3.7 (105–114) μm wide), with four equidistant papillae (two sub-dorsal and two sub-ventral) (Figure 10A, C). Oral aperture triangular under SEM, bordered by three lips with jaws between them, carrying radial teeth (Figure 10B). Pharynx short and powerful, with a robust corpus (288 ± 18.1 (267–311) μm long). Brown arcade cells surrounding the corpus in its distal part. Isthmus slightly delineated. Basal bulb stout (122 ± 8.3 (110–128) μm height x 139 ± 8.2 (129–147) μm wide). Nerve ring encircling pharyngeal corpus ca. its midpoint. Testis single, well-developed, filled with rounded spermatocytes. Tail short, conoid, ending in a fine point. Bursa absent. Spicules ventrally arcuate, almost isometric and isomorphic (Figure 9). Twenty-two copulatory papillae present. Pre-cloacal papillae arranged as seven pairs, four pairs postcloacal (Figure 10D–F).

Figure 9. Rhigonema disparovis Van Waerebeke Reference Van Waerebeke1991. LM micrographs of female and male. A: Entire female; B: Female pharyngeal region; C: Male tail region; D, E: Detail of vulval region showing eggs; F: Female tail. (Scale bars: A = 1000 μm; B–F = 100 μm)

Figure 10. Rhigonema disparovis Van Waerebeke Reference Van Waerebeke1991. SEM micrographs of male. A: Male lip region; B: En face view; C: Detail of microtrichs; D–F: Male tail showing pre- and post-cloacal papillae. (Scale bars: A = 15 μm; B = 10 μm; C = 5 μm; D, E = 100 μm; F = 20 μm)
Female. Body slightly larger than male. Morphology almost identical to that of male except for sexual characters. Ovaries opposed, often containing small oocytes; seminal receptacle containing small, rounded spermatozoa (diameter about 5 μm). Vagina anteriorly directed; vaginal diverticulum absent. Ovejector structure conforming to Type 1 (Adamson Reference Adamson1987), 352 ± 38.0 (300–391) μm long. Reduced vaginal chamber (no vaginal sac). Eggs ovoid with smooth shell, measuring 169 (159–196) x 110 (101–117) μm. Tail short, conoid.
Remarks
Morphology and morphometrics of the Nigerian specimens (Table 2) agree with those of the type population of this species and other reports from Ivory Coast (Van Waerebeke Reference Van Waerebeke1991; Hunt Reference Hunt2002). The main differences between the Nigerian population of R. disparovis and the type population are a slightly longer male body length ((5456–5962) vs (4080–5360)) μm, a slightly lower V ratio ((54.0–56.3) vs (55.4–60.7)), a longer pharynx length in females and males ((390–499, 406–452) vs (338–402, 332–70) μm, respectively); and a spicule length ((277–384) vs (389–458) μm). These small morphometrics differences may be due to geographical intraspecific variability. To our knowledge, this is the first report of this species from Nigeria.
Molecular characterization
Rhigonematids parasitizing millipedes from Nigeria were molecularly characterized by sequences of three ribosomal genes: the D2-D3 segment of 28S, ITS and partial 18S rDNA, and the mitochondrial gene COI (Table 1). The amplification of these regions yielded single fragments of approximately 900, 1200, 1600, and 700 bp, respectively, based on gel electrophoresis. Seventeen D2-D3 of 28S rDNA sequences from 693 to 747 bp (OQ627375–OQ627391), ten ITS rDNA sequences from 947 to 1203 bp (OQ633077–OQ633086), fourteen 18S rDNA sequences from 1537 to 1689 bp (OQ629072–OQ629085), and twelve COI sequences from 618 to 638 bp (OQ625295– OQ625307) were generated for the Nigerian rhigonematids (Table 1). Overall intraspecific variation was 0–1 nucleotides and 0 indel for D2–D3, 0–4 nucleotides and 0–2 indels for ITS, 0–27 nucleotides and 0–1 indel for 18S, and 0–21 nucleotides and 0 indels for COI. The D2–D3 segment of 28S rDNA of Brumptaemilius sp. was 96.8% similar to Brumptaemilius justini (JX999732) and 91.7% similar to Insulanema longispiculum (JX436471); the D2–D3 of G. gabonensis was 91.5% similar to Cattiena fansipanis (JX436470) and 91.0% similar to Insulanema longispiculum (JX436471); the D2–D3 of O. pachnephorus was 98.4% similar to Obainia sp. SVM-2017 (KU561100) and 93.5% similar to Rhigonema sinense (ON936080); the D2–D3 of R. disparovis was 85.7% similar to Rhigonema sp. (JX155275) and 86.5% similar to Rhigonema naylae (KX844643) and Rhigonema sinense (ON936086); and the unidentified juvenile nominated as Carnoyidae sp. 1 was 97.2% similar to Brumptaemilius sp. (OQ627375–OQ627382), 95.2% similar to Brumptaemilius justini (JX999732) and 90.8% similar to Insulanema longispiculum (JX436471). ITS rDNA of Brumptaemilius sp. was quite different from other ITS rhigonematids sequences in NCBI, with R. sinense (ON936112) being 77.4% similar but with a low sequence coverage only 55%; the ITS of G. gabonensis was 77.4% similar to R. sinense (ON936109) but with a low sequence coverage of only 45%; the ITS of O. pachnephorus was 80.7% similar to R. sinense (ON936107); and the ITS of R. disparovis was 79.6% similar to R. sinense (ON936109) but with a low sequence coverage of only 65%. The 18S rDNA of Brumptaemilius sp. was highly similar to several rhigonematids, including 99.9% similar to B. justini (JX999733) and 99.4% similar to Cattiena trachelomegali (JX982117); the 18S of G. gabonensis was 98.0% similar to B. justini (AF036589) and 97.5% similar to Heth tuxtlensis (KY857883) and other rhigonematids; the 18S of O. pachnephorus was 99.2% similar to Obainia sp. SVM-2017 (KU561101) and 99.0% similar to Rhigonema ingens (JX069475) and other rhigonematids; the 18S of R. disparovis was 98.1% similar to Rhigonema ingens (JX069475) and 97.5% similar to Xystrognathus phrissus (JX101957); and the 18S of the unidentified juvenile nominated as Carnoyidae sp. 1 was 99.9% similar to Brumptaemilius justini (JX999733), 99.8% similar to Brumptaemilius sp. (OQ629072–OQ629079), and 99.4% similar to Cattiena trachelomegali (JX982117). The COI of Brumptaemilius sp. was not found to be similar to other rhigonematids or other nematodes except for an 82.1% similarity to Litoditis aff. marina (KR815453); the COI of G. gabonensis was not similar to other rhigonematids or other nematodes except for an 82.0% similarity to Ancylostoma tubaeforme (KY070315); the COI of O. pachnephorus was 80.7% similar to R. sinense (ON935729) and 78.3% similar to R. naylae (OP113820); the COI of R. disparovis was not similar to other rhigonematids and was 77.6% similar to Cruznema tripartitum (OM234676) and 76.9% similar to Litoditis aff. marina (KR815451); and the COI of the unidentified juvenile nominated as Carnoyidae sp. 1 was 82.1% similar to Oesophagostomum sp. (MK282869) but showed a low similarity with other rhigonematids.
Phylogenetic relationships of rhigonematids from Nigeria with other Rhigonematina spp
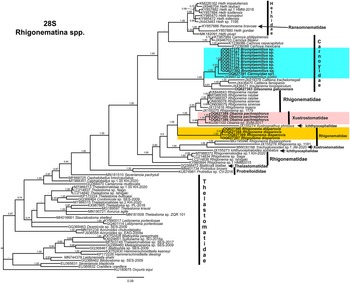
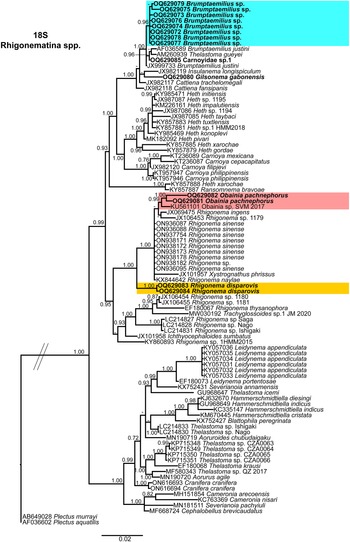
Phylogenetic relationships among rhigonematids species from Nigeria, as inferred from analyses of D2–D3 expansion domains of 28S, ITS, the partial 18S rDNA, and the partial COI mtDNA gene sequences using BI are shown in figures 11, 12, 13, and 14, respectively. The phylogenetic trees generated with the ribosomal and mitochondrial DNA markers included 88, 28, 93, and 30 sequences, and their alignment had 740, 1039, 1647, and 674 characters, respectively. In the 50% majority rule consensus, the D2–D3 of 28S rDNA BI tree of rhigonematids from Nigeria clearly separated the five identified species (Figure 11). Brumptaemilius sp. and the unidentified Carnoyidae sp.1 clustered with Brumptaemilius justini in a well-supported clade (PP = 1.00), and all of them clustered with other Carnoyidae species in a well-supported clade (PP = 1.00), including G. gabonensis, Insulanema longispiculum, Cattiena fansipanis, and Cattiena trachelomegali (Figure 11). Obainia pachnephorus clustered with Obainia sp. SVM-2017 in a well-supported clade (PP = 0.99), and R. disparovis clustered in a separate subclade from other Rhigonema species (Figure 11).

Figure 11. Phylogenetic relationships of rhigonematids from Nigeria with Rhigonematina species. Bayesian 50% majority rule consensus tree as inferred from D2 and D3 expansion domains of 28S rDNA sequence alignment under the GTR + I + G model (−lnL = 14186.47006; AIC = 28740.940120; freqA = 0.1926; freqC = 0.2065; freqG = 0.76; freqT = 0.2633; R(a) = 0.991; R(b) = 5.157; R(c) = 1.7505; R(d) = 0.780; R(e) = 7.8331; R(f) = 1.0000; Pinva = 0.2650; and Shape = 0.7550). Posterior probabilities greater than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and the coloured box indicates clade association of the new species. Scale bar = expected changes per site.

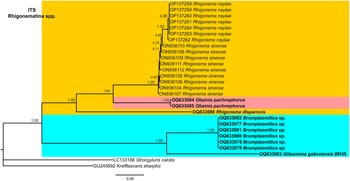
Figure 12. Phylogenetic relationships of rhigonematids from Nigeria with Rhigonematina species. Bayesian 50% majority rule consensus tree as inferred from ITS rDNA sequence alignment under the GTR + I + G model (−lnL = 5851.86800; AIC = 11831.736000; freqA = 0.2374; freqC = 0.2186; freqG = 0.2678; freqT = 0.2762; R(a) = 1.2073; R(b) = 4.0706; R(c) = 2.2937; R(d) = 1.0049; R(e) = 4.7765; R(f) = 1.0000; Pinva = 0.2950; and Shape = 1.5100). Posterior probabilities greater than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and the coloured box indicates clade association of the new species. Scale bar = expected changes per site.

Figure 13. Phylogenetic relationships of rhigonematids from Nigeria with Rhigonematina species. Bayesian 50% majority rule consensus tree as inferred from 18S rDNA sequence alignment under the SYM + I + G model (−lnL = 7588.03030; AIC = 15558.060600; freqA = 0.2500; freqC = 0.2500; freqG = 0.2500; freqT = 0.2500; R(a) = 1.2300 R(b) = 3.1348; R(c) = 2.3869; R(d) = 0.6116; R(e) = 5.5863; R(f) = 1.0000; Pinva = 0.5730; and Shape = 0.6200). Posterior probabilities greater than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and the coloured box indicates clade association of the new species. Scale bar = expected changes per site.

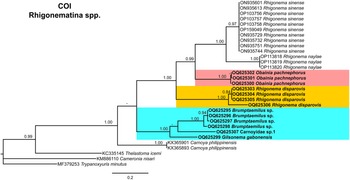
Figure 14. Phylogenetic relationships of rhigonematids from Nigeria with Rhigonematina species. Bayesian 50% majority-rule consensus trees as inferred from cytochrome c oxidase subunit I (COI) mtDNA gene sequence alignments under the TVM + I + G model (−lnL = 4679.81712; AIC = 9493.634240; freqA = 0.2286; freqC = 0.073435; freqG = 0.2200; freqT = 0.4780; R(a) = 0.2662; R(b) = 7.5805; R(c) = 0.9593; R(d) = 3.6092; R(e) = 7.5805; R(f) = 1.0000; Pinva = 0.1580; and Shape = 0.3880). Posterior probabilities greater than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold, and the coloured box indicates clade association of the new species. Scale bar = expected changes per site.
The ITS tree was constructed with rhigonematids sequences from Nigeria and the very few suitable sequences of rhigonematids in NCBI (Figure 12). Brumptaemilius sp. and G. gabonensis clustered together in a well-supported clade (PP = 1.00). The ITS tree confirmed the separate clustering of R. disparovis with other Rhigonema spp. (R. naylae and R. sinense), and O. pachnephorus clustered together with R. naylae and R. sinense (Figure 12).
In the 18S rDNA BI tree, all three Carnoyidae species from Nigeria (Brumptaemilius sp., G. gabonensis, and Carnoyidae sp. 1) clustered together in a well-supported clade (PP = 1.00) with other Carnoyidae species (B. justini, I. longispiculum, C. fansipanis, C. trachelomegali) and thelastomatid species (Thelastoma gueyei) (Figure 13). Obainia pachnephorus and R. disparovis clustered together with other Rhigonema species (R. ingens, R. naylae, R. sinense, Rhigonema sp.) and Xystrognathus phrissus in a well-supported clade (PP = 1.00) (Figure 13).
Finally, the phylogenetic relationships of Nigerian rhigonematids with other species, using COI gene sequences, clustered O. pachnephorus with R. disparovis and R. sinense and R. naylae in three separate well-supported (PP = 1.00) subclades of a well-supported clade (Figure 14). Brumptaemilius sp. and G. gabonensis clustered together in a well-supported clade (PP = 1.00), but the relationship with Carnoyidae sp. 1 was not well resolved (Figure 14). The two only thelastomatid species with COI available (Thelastoma icemi and Cameronia nisari) are clearly separated from all the others and more data are needed to confirm their position as outgroups together with the oxyurid Trypanoxyuris minutus (Figure 14).
Discussion
Accurate species identification of rhigonematids parasitizing millipedes is often problematic due to many species being poorly described morphologically owing to a combination of a great phenotypic similarity within females of the same genus and a scarcity of accurate detail of the male posterior region (Van Waerebeke Reference Van Waerebeke1984; Hunt Reference Hunt2001a). In addition, few integrative taxonomic studies (including morphometric and molecular characterizations) have been done in these nematodes, which is something that must be considered a keystone in future. The main objective of this study was to identify rhigonematids from Nigeria using integrative taxonomical approaches, as well as to clarify and provide new insights into the phylogenetic relationships within these nematodes. All the provided results confirmed that the studied Nigerian rhigonematids belong to five species clearly separated by morphological traits and molecular markers. To our knowledge, this is the first report of G. gabonensis, O. pachnephorus, and R. disparovis in Nigeria, and it represents the first molecular characterization of these species.
Ribosomal and mitochondrial markers (D2–D3 expansion domains of the 28S and ITS rRNA, and the mtDNA gene COI) are important tools for accurate identification of species in the Phylum Nematoda and remain essential for accurate diagnosis and phylogenetic relationships of rhigonematids (Malysheva et al. Reference Malysheva, Van Luc and Spiridonov2012; Kim et al. Reference Kim, Kim, Cho, Min, Park, Carreno, Nadler and Park2014; Malysheva et al. Reference Malysheva, Mohagan and Spiridonov2015; Mejia-Madrid Reference Mejia-Madrid2018; Zhang et al. Reference Zhang, Wang, Hasegawa, Hasegawa, Nagae, Li LW and Li2022). However, the low availability of ITS rRNA sequences makes it difficult to infer phylogenetic relationships with this marker. Phylogenetic analyses based on 28S and 18S ribosomal genes resulted in a general consensus of species phylogenetic positions for the majority of species and was congruent with those given by previous phylogenetic analyses (Malysheva et al. Reference Malysheva, Van Luc and Spiridonov2012; Kim et al. Reference Kim, Kim, Cho, Min, Park, Carreno, Nadler and Park2014; Malysheva et al. Reference Malysheva, Mohagan and Spiridonov2015; Mejia-Madrid Reference Mejia-Madrid2018; Zhang et al. Reference Zhang, Wang, Hasegawa, Hasegawa, Nagae, Li LW and Li2022). Our results on 28S and 18S rRNA genes suggest that genera within Ransomnematoidea (Ransomnema, Heth, Carnoya, Brumptaemilius, Cattiena, Insulanema, Gilsonema) and Rhigonematoidea (Rhigonema, Obainia, Xystrognathus, Trachyglossoides, Ichthyocephaloides) clustered closer than could be expected in view of their morphological differences, these groups having been lumped together mainly on host association, although it would appear from the molecular data that they are actually closely linked (Figures 11, 13). De Ley and Blaxter (Reference De Ley, Blaxter and Lee2002) questioned the validity of classifying Rhigonematoidea and Ransomnematoidea as sister taxa based on sparse information, but our results based on ribosomal genes (28S and 18S) support the sister relationships of both subfamilies, and to a lesser extent with the mitochondrial gene (COI). Mejia-Madrid (Reference Mejia-Madrid2018) reported that Ransomnema bravoae represents a rogue taxon appearing in an unresolved position within Rhigonematoidea in 18S rRNA phylogeny, whereas in the combined 18S and 28S rRNA analyses, it appears more closely related to the Carnoyidae and Hethidae; however, our 18S and 28S rRNA analyses showed high PP values (Figures 11, 13). In any case, as suggested by Mejia-Madrid (Reference Mejia-Madrid2018), the inclusion of more genera and species of rhigonematids for further phylogenetic analyses is required in order to recover more detailed relationships within the major clades that comprise the Suborder Rhigonematina. In our analyses, the genus Rhigonema (one of the more studied from a molecular point of view) appears polyphyletic (fFigures 11, 13). Although genera within Thelastomatoidea (Leidynema, Severianoia, Hammerschmidtiella, Thelastoma, Aorurus, Cranifera, Cameronia, Cephalobellus) clustered together in a well-supported clade in the 18S rRNA tree (Figure 13), they were not well-resolved in the 28S rRNA tree (Figure 11). Finally, although relationships on mitochondrial COI are congruent with those of ribosomal genes, they are not conclusive due to the scarcity of available sequences of this gene in NCBI. However, COI is also suggested as an excellent molecular marker for species separation and identification as reported by Malysheva et al. (Reference Malysheva, Efeykin and Teterina2016) and Zhang et al. (Reference Zhang, Wang, Hasegawa, Hasegawa, Nagae, Li LW and Li2022).
All these data confirm previous hypotheses that Suborder Rhigonematina may be considered as polyphyletic since two superfamilies Ransomnematoidea and Rhigonematoidea formed independent clades in ribosomal and mitochondrial genes (Nadler et al. Reference Nadler, Carreno, Mejia-Madrid, Ullberg, Pagan, Houston and Hugot2007; Malysheva et al. Reference Malysheva, Mohagan and Spiridonov2015).
In summary, the present study confirms the usefulness of applying an integrative approach based on a combination of morphometric and morphological traits and genotyping rRNA and mtDNA markers for correct species discriminating among rhigonematids species and their phylogenetic relationships, suggesting the need for continuing nematode surveys of millipede parasites in other areas in order to complete the unexplored biodiversity of these nematodes.
Acknowledgements
The authors thank J. Martín Barbarroja and G. León Ropero for their excellent technical contributions. SEM pictures were obtained with the assistance of technical staff and the equipment of Centro de Instrumentación Científico-Técnica (CICT), University of Jaén.
Competing interest
The authors have no competing interests.
Ethical standard
The result of this work has not been published previously and is not under consideration elsewhere.
Ethical approval
The conducted research is not either related to human or animals use.