INTRODUCTION
Pneumonia remains a major global public health problem, and an important focus for scientific study [Reference Rudan1]. Identifying causative agents can be a challenge because the lung itself is rarely sampled and culturing of blood or respiratory secretions can be insensitive or difficult to interpret [Reference Mizgerd2]. There is still a paucity of data from low- and middle-income countries, particularly in tropical and subtropical areas, and it is likely that the causative agents, their seasonal and age distributions, and other aspects of pneumonia epidemiology differ in these settings and require tailored prevention and control strategies.
Beginning in 2002, the Thailand Ministry of Public Health and the U.S. Centers for Disease Control and Prevention (CDC) combined resources to establish active pneumonia surveillance for persons of all ages in two Thai provinces [Reference Olsen3]. Global events such the SARS outbreak, avian influenza A (H5N1) and 2009 influenza A (H1N1) infections in humans fuelled interest in and funding for influenza and other viruses that previously received little attention [4–Reference Dawood8]. The threat of emerging pathogens presenting as pneumonia emphasized the importance of effective epidemiological and clinical surveillance integrated with reliable laboratory diagnostics. Here we present results for the first 30 months of active pneumonia surveillance in Thailand, focusing on the incidence of viral and atypical bacterial pathogens.
METHODS
Setting
Thailand is a middle-income country with a gross national income of U.S. $2990 per capita, a low infant mortality rate (7 deaths/1000 live births) and an average life expectancy of 70 years [9]. It has a tropical climate with a mid-May to September monsoon season. An enhanced surveillance system to actively identify prospective cases of pneumonia requiring hospitalization began in Sa Kaeo province in 2002 and was expanded to Nakhon Phanom province in 2003. Sa Kaeo (population 514 065 in 2003) is in eastern Thailand and borders Cambodia, and Nakhon Phanom (population 730 659 in 2005) is in northeast Thailand and borders Laos. The combined population of these two provinces accounts for ~2% of the total population of Thailand.
Study population
Pneumonia surveillance is described in detail elsewhere [Reference Olsen3]. Briefly, surveillance officers actively identified patients hospitalized at any of the eight hospitals (one provincial, six district and one military) in Sa Kaeo province and any of the 12 hospitals (one provincial, ten district and one military) in Nakhon Phanom province. There are no private hospitals in these two provinces and through community surveys we know that the hospitalized surveillance system captures 58–80% of all pneumonias in these two provinces [Reference Chamany10, Reference Jordan11]. Surveillance identified patients with active infection (either reported fever, reported chills, measured temperature >38·2°C or <35·5°C, or an abnormal white blood cell count or differential), and lower respiratory tract disease (either abnormal breath sounds, documented tachypnoea, observed cough, sputum production, or dyspnoea). Basic demographic, clinical and laboratory findings were collected on a standard surveillance form. Patients who had a chest radiograph performed within 48 h of admission were approached for enrolment into the aetiological study. Chest radiographs were digitized and sent to Bangkok to be interpreted by a panel of radiologists using WHO criteria as previously described [Reference Javadi12]. Here we analysed data from a period when complete viral aetiological information was available – between 1 September 2003 and 31 August 2005 (2 years for Sa Kaeo) and between 1 January and 31 December 2005 (1 year for Nakhon Phanom). This study was approved by a CDC Institutional Review Board (protocol no. 3754) and the Ethical Review Board of the Thailand Ministry of Public Health.
Specimen collection
Consenting patients provided nasopharyngeal swab, urine, and acute and convalescent serum specimens [Reference Simmerman13]. Briefly, within 48 h of admission and after informed consent, one Dacron® swab was placed into one nostril to the nasopharynx and rotated once to collect epithelial tissue and absorb secretions. Swabs were placed in viral transport media, refrigerated at 2–8°C for up to 48 h, and then the media was aliquoted and stored at −70°C. A blood sample was collected at the time of enrolment and again 3 weeks later (5 ml of blood from children aged <3 years and 10 ml for all others); serum was aliquoted and stored at −70°C in Thailand and −20°C after transfer to the USA. A 10-ml urine sample was collected and refrigerated at 2–8°C at each hospital until it was transported to the provincial hospitals each day and frozen at −70°C. Specimens were transported on dry ice once weekly to the laboratory in Bangkok for specimen processing and shipping to CDC in Atlanta, GA, USA.
Laboratory tests
The summary of laboratory testing methods is shown in Table 1. Methods used include currently available diagnostic assays for use in the hospital setting in rural Thailand, commercially available assays, as well as research assays developed by CDC.
Table 1. Laboratory methods used to identify respiratory pathogens

NP, Nasophayrngeal; PCR, polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; RT–PCR, reverse transcriptase–polymerase chain reaction.
Hospital tests
Testing for Mycobacterium tuberculosis was performed by acid fast bacilli (AFB) smear of patient sputum in the hospital laboratory at the discretion of the attending physician. HIV testing was performed using the local hospital's routine method which included ELISA, Western blot, radioimmunoprecipitation, immunofluorescence assay (IFA) or gelatin particle agglutination. HIV testing was performed at the discretion of the doctor until January 2005, when we began to actively approach patients for their consent. Positive tests were reported to the patient's physician and counselling was provided by the hospital according to established local practices. HIV-positive patients were referred into the existing Thailand health system for management. Hospitals did not routinely perform blood cultures.
Rapid antigen tests
BinaxNOW® rapid immunochromatographic assays (Binax Inc., USA) were used to detect Streptococcus pneumoniae and Legionella pneumophila serogroup 1 antigen in patients' urine. Laboratory technicians followed the manufacturer's instructions. Only patients aged ⩾18 years were tested for S. pneumoniae and L. pneumophila.
Viral cultures
Culture for influenza viruses, respiratory syncytial virus (RSV), adenovirus, parainfluenza viruses and metapneumovirus was performed at the Thailand National Institute of Health laboratories. The supernatant from nasopharyngeal swabs was inoculated in Madin–Darby canine kidney cells (MDCK), Hep-2 human epithelial cells (HEp-2) and Macaca mulatta (rhesus monkey) kidney cells (LLC-MK2). Cells were examined daily for cytopathic effect (CPE). Cells showing CPE were tested immediately by commercial IFA (Chemicon International Inc., USA) for the five groups of viruses being investigated. Cultures showing no CPE after 7–10 days' incubation were passed into fresh cells and monitored as above. If no CPE developed after the second passage, all cells were harvested and reconfirmed by IFA using pooled monoclonal antibodies; positives were further tested with specific monoclonal antibodies.
Polymerase chain reaction (PCR) assays
PCR was performed only on nasopharyngeal samples. Reverse transcriptase–polymerase chain reaction (RT–PCR) assays were developed at CDC to detect RSV, parainfluenza viruses 1, 2 and 3 (HPIV1, 2, 3), influenza viruses types A and B, rhinoviruses, metapneumovirus, adenoviruses, bocavirus and coronaviruses. Samples collected during the 2 years in Sa Kaeo and first 6 months in Nakhon Phanom were tested by GeneScan RT–PCR assays as previously described [Reference Erdman14]. Samples collected during the last 6 months in Nakhon Phanom were tested by newly developed real-time RT–PCR assays. In direct comparisons with GenScan RT–PCR assays, the real-time RT–PCR assays showed comparable sensitivity with serially diluted viral nucleic acid and gave identical results with a sample of GenScan RT–PCR positive and negative study samples (D. Erdman, CDC, personal communication). To identify rhinoviruses, picornavirus-positive samples were subjected to a second round of semi-nested PCR using primers specific for rhinovirus [Reference Schrag15]. PCR assays were recently developed for bocavirus [Reference Lu16] and coronaviruses 229E, OC43, HKU1, and NL63 [Reference Dare17]. Real-time PCR assays for the detection of Mycoplasma pneumoniae and Chlamydia pneumoniae were developed at CDC [Reference Tondella18, Reference Zhang19].
Serology
Chlamydia pneumoniae serology (IgG and IgM) was performed using a microimmunofluorescence assay (Focus Diagnostics, USA). Legionella longbeachae combined IgG, IgA and IgM titres were measured using an indirect IFA (only patients aged ⩾18 years) [Reference Wilkinson20]. Mycoplasma pneumoniae serology (IgG and IgM) was performed using a semi-quantitative ELISA (Remel, USA). Coxiella burnetii serology (IgM and IgG) was measured by ELISAs [Reference Field21, Reference Field22]. Influenza serology was performed with the haemagglutinin inhibition test. RSV, HPIV 1, 2 and 3, adenovirus and metapneumovirus IgG antibodies were tested by ELISA [Reference Dowell23–Reference Marx25].
Analysis and definitions
Mid-year population estimates for Sa Kaeo and Nakhon Phanom were obtained using data from the National Economic and Social Development Board of Thailand [26]. Crude incidence was calculated as a population-weighted average for the 6 months when both sites had data (January–July 2005). We adjusted the incidence for enrolment by multiplying the number of patients eligible by the proportion that tested positive for a given pathogen in those patients enrolled. Monthly unadjusted incidence is shown only for viruses with a monthly incidence above 1/100 000 (excludes parainfluenzaviruses 2 and 3, and the coronaviruses). Recurrent pneumonia was defined as two episodes of pneumonia separated by at least 14 days. Viral infections were determined by a positive RT–PCR result, positive viral culture or fourfold rise in antibody titre. HIV was considered positive if any of the hospital tests were reported as positive. Legionella pneumophila infections were determined by urine antigen test positivity and L. longbeachae by a fourfold rise in antibodies or PCR positivity. C. pneumoniae infections were determined by a positive PCR or serology result (fourfold rise in IgG titre and/or IgM ⩾16) [Reference Dowell27]. M. pneumoniae infections were defined as a seroconversion from negative in acute serum to positive in convalescent serum, or PCR positivity. Tuberculosis was defined using the WHO definition of at least two AFB sputum smear-positive results or one positive smear with an abnormal chest radiograph [28]. To distinguish recognized pathogens from agents with an unclear pathogenic role in pneumonia, we defined a category named ‘recognized pneumonia aetiologies’ which included any of the respiratory pathogens except bocavirus, coronaviruses and rhinoviruses. Proportions were compared using χ2 and differences in means using the t test. All analyses were performed using SPSS version 15.0 (SPSS Inc., USA).
RESULTS
Between September 2003 and December 2005, surveillance officers identified 15 403 patient admissions with active infection and lower respiratory tract disease. Of these, 8351 (54%) received a chest radiograph and were approached for consent and enrolment into the aetiological study; 3910 (46·8%) were enrolled. Enrolled patients were more likely to be older (median age 32 vs. 24 years, P=0·001) than those who were not enrolled [Reference Olsen3, Reference Phares29].
A chest radiograph was available for reading by the radiologist panel for 2444 (62·5%) of pneumonia episodes. Of those read, 1730 radiographs in 1689 patients (70·8%) had evidence of pneumonia; 1293 (74·7%) of the abnormal radiographs showed interstitial infiltrates, 1047 (60·5%) showed alveolar infiltrates (675 showed evidence of both interstitial and alveolar infiltrates) and 65 (3·8%) had other evidence of pneumonia. Of the 1730 episodes of radiographically confirmed pneumonia, 599 (34·6%) were in children aged <5 years and 1013 (58·6%) were in males. The median length of hospital stay was 5 days (range 1–139). A total of 114 (6·6%) patients had evidence of HIV infection [34/695 (4·9%) patients were tested as part of the study and an additional 80 had evidence of previous testing or clinical evidence in the medical chart]. Of the 114 with any evidence of having HIV, four were aged <5 years, five were aged 5–17, 92 were aged 18–49 and 13 were aged ⩾50 years.
Aetiological agents and incidence
Of the 1730 episodes of radiographically confirmed pneumonia, 231 (13·4%) had a bacterial agent identified, 746 (43·1%) had a viral agent identified, and 176 (10·2%) had mixed infections (Table 2). Of pneumonia episodes in children aged <5 years, 2·2% had a bacterial agent identified, 75·6% a viral agent and 15·2% had mixed infections. Children aged <5 years had a higher proportion of pathogens identified compared to all other ages; the difference was statistically significant (P<0·001).
Table 2. Number and percent of respiratory pathogens by age group detected in specimens from 30 months of pneumonia surveillance in Thailand, 1 September 2003 to 31 December 2005. Denominators are listed in cells when testing was not complete for the 30 months

CXR, Chest X-ray; n.t., not tested.
* Exluding bocavirus.
† Excluding bocavirus, coronviruses and rhinoviruses.
Crude and adjusted pathogen-specific incidence in radiographically confirmed pneumonia is shown in Table 3 [Supplementary Table S1 (available online) presents incidence in all patients enrolled]. Of children aged <5 years, RSV had the highest adjusted weighted average annual incidence (all values are per 100 000) (417·1), followed by rhinoviruses (240·9), bocavirus (114·6), and influenza A (90·2). In persons aged 5–49 years, rhinovirus incidence was highest (7·2), followed by M. tuberculosis (5·7), influenza A (3·9) and RSV (3·2). Of persons aged ⩾50 years, influenza A incidence was the highest (38·8) followed by rhinoviruses (37·1), S. pneumoniae (34·7) and M. tuberculosis (30·5).
Table 3. Population-weighted crude and adjusted annual incidence of radiographically confirmed pneumonia per 100 000 by age group

n.t., Not tested.
* Adjusted for enrolment.
Mixed viral infections
Overall, 5·0% (87/1730) of all episodes of radiographically confirmed pneumonia (11·7% of episodes in children aged <5 years) had more than one virus identified (Table 4). A rhinovirus was identified in 54·0% (47/97) of all mixed infections.
Table 4. Mixed viral infections in episodes of hospitalized, radiographically confirmed pneumonia by age group (bocavirus is excluded)
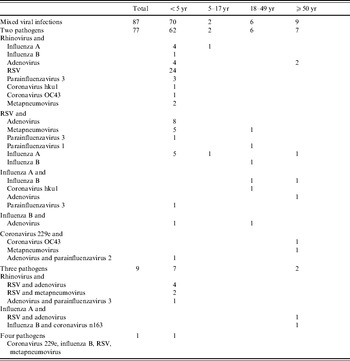
RSV, Respiratory syncytial virus.
Seasonality
The monthly incidence of radiographically confirmed pneumonia peaked twice each year, in January–March and June–September (Fig. 1). The peak in January–March included pneumonia patients with parainfluenza virus 3 and bocavirus. In contrast, the June–September peak included pneumonia patients with RSV, metapneumovirus and adenovirus. Influenza viruses A and B and bocavirus had peaks in both pneumonia seasons. June–August 2005 had the highest incidence of RSV infection; the all-age monthly incidence peaked in July at 9·0/100 000. Rhinoviruses were identified throughout the study period with no distinct seasonality.

Fig. 1. Averaged monthly incidence of radiographically confirmed pneumonia (shaded in grey) and (a) respiratory syncytial virus, (b) influenza viruses A and B, (c) metapneumovirus and parainfluenza virus 3, (d) bocavirus, adenovirus and rhinoviruses.
Pneumonia mortality
Overall, 66 (3·8%) pneumonia episodes were fatal, but this varied by year and site [28/478 (5·9%) in Sa Kaeo year 1, 29/512 (5·7%) in Sa Kaeo year 2 and 9/740 (1·2%) in Nakhon Phanom]. Of the 66 pneumonia deaths, 40 (60·6%) were male and the median age was 69 years (range 4 months to 90 years). Three deaths (4·5%) were in children aged <5 years. Fifteen (22·7%) persons had evidence of HIV infection. Fifteen (22·7%) of the patients who died had at least one respiratory pathogen identified: four had evidence of rhinoviruses (one also with HIV), five had evidence of tuberculosis (two also with HIV, one with coronaviruses 229 and OC43), four of pneumococcus (one also with HIV) and two of influenza A (one also with C. pneumoniae). There was no identified aetiology in the three deaths in young children. Of patients who died, the median length of hospital stay was 7 days (range 1–43). One elderly patient with influenza who died had an extended length of stay (139 days) due to several pre-existing medical comorbidities [Reference Simmerman13].
DISCUSSION
This pneumonia study includes patients of all ages from a well-described population and integrates epidemiological and clinical surveillance with thorough viral laboratory diagnostics to expand the understanding of the viral causes of pneumonia in a middle-income country in a tropical area. It also adds to valuable information about pneumonia derived largely from wealthy temperate countries in studies over the past 30 years [Reference Filice30–Reference Jennings37]. The population-based design, capturing all hospitalized pneumonia cases in the 1·2 million population of the two provinces allows for rates of disease to be calculated and compared, a feature of only a few previous studies [Reference Filice30, Reference Marston32, Reference Jackson36, Reference Jennings37]. Overall, the study provides data that may help to guide empirical treatment, target pneumonia prevention strategies and direct future research in Thailand.
Viral pathogens, especially RSV, caused a substantial proportion of pneumonias in young children, a finding that is consistent with other studies around the world [Reference Weber, Mulholland and Greenwood38, Reference Stensballe, Devasundaram and Simoes39]. The adjusted incidence of RSV-associated pneumonia in children was 417·1/100 000. Viruses were also important in hospitalized adult pneumonias, perhaps an under-appreciated finding [Reference Dowell23, Reference Marx25, Reference Marston32, Reference Jennings37]; 8·7% of patients aged ⩾50 years had influenza A infection and 2·9% had RSV infection. Improved prevention strategies, such as vaccination or improved hand hygiene, to combat common viruses such as influenza and RSV might greatly reduce hospitalized pneumonia in Thailand.
In Thailand, there appears to be two distinct pneumonia seasons with different pathogens contributing to each. Viral activity is known to vary from year to year. For example, in the USA, RSV has an annual peak but year-to-year and geographic variability is quite great [Reference Mullins40]; and the alternating year pattern of parainfluenza virus activity is well recognized [Reference Fry41]. In the tropics, RSV has been associated with the rainy season [Reference Weber, Mulholland and Greenwood38], a finding consistent with the pattern we found in Thailand. In the current study, we believe surveillance artifact (i.e. a nurse strike in one province) may have contributed to the lower RSV peak observed in 2004. Several more years of data will help distinguish recurrent seasonal patterns from temporary aberrations or outbreaks. Seasonality is important to describe since it has implications for timing of preventive measures; the circulation of influenza A and B almost year round would complicate any vaccine strategy.
Incidence is important for understanding the burden of disease, and it is an important tool for policy makers. In this study, patients signed informed consent before undergoing the extensive diagnostic workup, and enrolment averaged 48% over the 30 months and was lowest in children. Furthermore, only 62·5% of those enrolled had a chest radiograph. To more accurately estimate incidence, we made adjustments (Table 3). Of note, many viral diseases present with pneumonia or another clinical syndrome (e.g. RSV can present as pneumonia or brochiolitis) so our incidence probably underestimates the true burden.
A large limitation of this study is the non-standard use of diagnostic assays to confirm bacterial causes of pneumonia. As a result, we are unable to describe the bacterial aetiology of childhood pneumonia in this setting. Adults aged ⩾50 years had an elevated incidence of tuberculosis and two vaccine-preventable diseases, influenza and pneumococcal pneumonia. The incidence of tuberculosis is likely to be a gross underestimate since AFB smears were used at the discretion of the physicians. In addition, atypical bacterial agents diagnosed by urine antigen, PCR assay or serology were also common in older adults with pneumonia. The adjusted incidence of C. pneumoniae was 25·5/100 000 in adults aged ⩾50 years. However, commercial serological tests for C. pneumoniae have not been well validated clinically and therefore sensitivity, specificity and predictive values for these tests are not accurately known. L. longbeachae was an interesting finding and deserves more study, as was the notable absence of L. pneumophilia.
Improved molecular diagnostics have radically changed our understanding of the role of viruses in respiratory disease. However, with improved sensitivity comes a greater challenge in data interpretation, namely how to interpret mixed viral infections. In this study, rhinoviruses were most frequently identified in mixed infections. We also found viral co-infections to be much more frequent in young children than in older children and adults, a finding that probably reflects the increased probability of young children to acquire viral infections. Clinical and epidemiological data, as well a control population, will be critical to proper interpretation of the identified association between selected viruses and pneumonia and to determining causality vs. carriage.
Pneumonia remains a leading infectious killer worldwide, and the increased attention in Asia and tropical countries, where viral respiratory diseases including SARS and H5N1 influenza viruses emerged, is warranted. Importantly, a large proportion of pneumonia is caused by known treatable or preventable infectious agents. At the local level information on the causes of pneumonia will allow clinicians to better understand pneumonia aetiology and make informed treatment decisions. In Thailand, molecular diagnostic capabilities (i.e. RT–PCR) are being added to the provincial hospital laboratories in these surveillance sites. At the national and regional level, these data will allow public health officials to make informed policy decisions on prevention strategies, such as Thailand's recent decision to publicly provide influenza vaccine to elderly persons. At the global level, comprehensive surveillance which integrates epidemiological, clinical and laboratory information in the context of capacity building and technology transfer will contribute to better preparedness against the next infectious disease threat.
ACKNOWLEDGEMENTS
We thank Peera Areerat and Denchai Sornkij for their senior leadership in the field; Sununta Henchaichon, Suchada Kaewchana, Prabda Prapasiri, Sathapana Naorat, Possawat Jorakate and Anek Kaewpan for technical assistance in the field; Julie Fisher, Julia Rhodes, Prasong Srisaengchai, Pongpun Sawatwong and Pornpak Khunatorn for assistance in data management; Sona Budhiraja, Vondguraus McClee, George Gallucci, Julita Yarwood and Henrietta Hall for assistance in the laboratory. Funding was provided by the Centers for Disease Control and Prevention and the Thailand Ministry of Public Health. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of CDC.
NOTE
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/hyg).
DECLARATION OF INTEREST
None.







